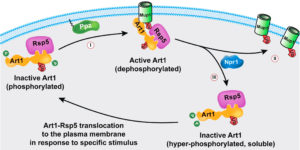
One mechanism that cells use to modulate the activity of plasma membrane proteins is to remove them from the surface through endocytosis. However, this mechanism requires that proteins to be removed are correctly identified so that needed membrane functions are not compromised. Ubiquitylation is one way that cells label plasma membrane proteins for endocytosis, though our understanding of how the key E3 ubiquitin ligases target the correct proteins is far from complete. This led Vanderbilt Basic Sciences investigator Jason MacGurn and his laboratory to study how the E3 ubiquitin ligase Rsp5 carries out this function in yeast cells. Previous research had revealed that ART (arrestin-related Rsp5 adaptor) proteins bind to Rsp5 and guide it to proteins for ubiquitylation. The MacGurn lab focused on the role of one of these proteins, Art1, in targeting Rsp5 to the yeast methionine transporter, Mup1. They began by expressing Mup1 fused to green fluorescent protein (Mup1-GFP) so that they could visualize its internalization through fluorescence microscopy. This enabled them to show that addition of methionine to the cells led to rapid uptake of Mup1-GFP from the plasma membrane followed by its sequestration into intracellular vacuoles. They also used expression of Art1-GFP to show that the internalization of Mup1 was preceded by translocation of Art1 to the plasma membrane, and they demonstrated that methionine-mediated Mup1 internalization did not occur in Art1 knockout cells. As activation of Art1 was known to occur through dephosphorylation of the protein, and because their prior work indicated a role for the Ppz phosphatases (Ppz1 and Ppz2) in ubiquitin homeostasis in yeast, the researchers hypothesized that these enzymes might be responsible for dephosphorylation and activation of Art1. Consistent with this hypothesis, methionine-mediated internalization of Mup1 did not occur in Ppz double knockout cells (Δppz1Δppz2). Expression of wild-type Ppz1 in Δppz1Δppz2 cells partially restored Mup1 internalization. However, mutations of Ppz1 that rendered it catalytically inactive or unable to bind to the plasma membrane eliminated its ability to restore methionine-mediated endocytosis of Mup1 in Δppz1Δppz2cells. Further studies revealed that methionine addition to yeast cells led to dephosphorylation of Art1 at Thr93, Thr245, and Thr795 and that this dephosphorylation did not occur in Δppz1Δppz2mutant cells. Expression of mutant Art1 proteins that mimicked permanent phosphorylation or prevented phosphorylation at these sites suggested that phosphorylation at Thr93 or Thr795 suppresses Art1 activation.Finally, the researchers showed that translocation of Art1 to the membrane in response to methionine was not affected by the Δppz1Δppz2double mutations, but the ability of Art1 to interact with Mup1 was suppressed. Together the results suggest that Ppz-mediated dephosphorylation of Art1 at Thr93 and Thr795 is required so that Art1 can bind to Mup1, thereby enabling Art1 to guide Rsp5 to Mup1 for ubiquitylation. Thework is published in the Journal of Cell Biology[S. Lee et al., (2019) J. Cell Biol., published January 4, DOI: 10.1083/jcb.201712144].