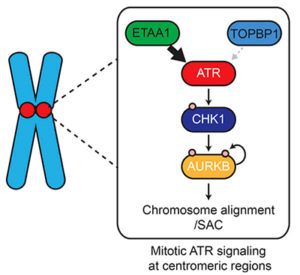
ATR (ataxia telangiectasia-mutated and Rad3-related) is a kinase well known for its role in the DNA damage and replication stress responses; however, it also is an important modulator of the cell cycle and mitosis. In mammalian cells, activation of ATR is mediated by two proteins, ETAA1 and TOPBP1, both of which associate with the kinase through very similar ATR-activation domains (AADs). However, ETAA1 and TOPBP1 interact differently with DNA and other proteins, suggesting that they play distinct roles in modulation of ATR function. To test this hypothesis, Vanderbilt Basic Sciences investigator David Cortez and his graduate student Thomas Bass used phosphoproteomics to identify protein targets of ETAA1- and TOPBP1-dependent ATR activity. To achieve this goal, they created HCT116 cell lines deficient in their ability to activate ATR via ETAA1 or TOPBP1 or both proteins. They found that when exposed to a replicative stress, all three cell lines exhibited defects in ATR-dependent signaling, and these defects were most pronounced in cells lacking both ETAA1 and TOPBP1 function. Increased levels of DNA damage, however, only occurred in TOPBP1-deficient cells, consistent with distinct roles for the two proteins. The investigators used SILAC (stable isotope labeling of amino acids in cell culture) and mass spectrometry-based phosphoproteomic analysis to identify and quantify proteins phosphorylated in response to ETAA1- or TOPBP1-dependent ATR activation in the cell lines. This approach characterized 285, 724, and 118 phosphosites (sites of phosphorylation) resulting from activation of ATR by ETAA1, TOPBP1, or both proteins, respectively. Using gene ontology (GO) analysis to determine major pathways associated with these proteins, the researchers concluded that TOPBP1-mediated activation of ATR is primarily important in the cellular response to DNA damage and replicative stress whereas ETAA1 controls ATR activity necessary for modulation of the cell cycle and mitosis under physiological conditions. Further studies using cells expressing Flag-tagged ETAA1 to enable isolation of the protein along with its intracellular binding partners demonstrated that ETAA1 interacts with numerous mitotic proteins, including proteins associated with the centromere and mitotic spindle. These findings were consistent with their phosphoproteomics data. The researchers also found that loss of ETAA1 but not TOPBP1 function led to a marked increase in chromosomal abnormalities and failure of correct chromosome separation during mitosis. Furthermore, cells lacking ETAA1-dependent ATR modulation failed to halt mitosis in the presence of spindle dysfunction caused by exposure to taxol. They concluded that this failure of the spindle assembly checkpoint was likely the result of the loss of signaling from Aurora B, a kinase that is activated by ATR via an intermediate kinase, CHK1. Together the results support the hypothesis that replication stress leads to ATR activation primarily through TOPBP1, whereas ETAA1 activates ATR under physiological conditions to modulate the cell cycle and mitosis. The work is published in the Journal of Cell Biology [T. E. Bass and D. Cortez, (2019) J. Cell Biol., published February 12, DOI: 10.1083/jcb.201810058].