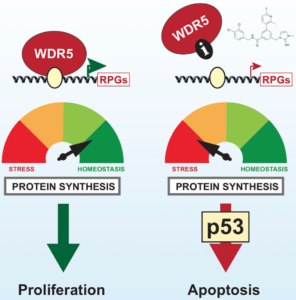
WDR5 serves as a scaffold for protein complexes containing epigenetic “writers” that catalyze histone modification reactions. Among the WDR5-dependent complexes are those encompassing MLL/SET-type histone methyltransferases (HMTs) responsible for di- and trimethylation of histone 3 at lysine-4 (H3K4). WDR5 contains a WIN (WDR interaction) site, which binds to an arginine-containing WIN motif on other proteins with which it interacts. Inhibitors of the WIN site-WIN motif interaction suppress the activity of MLL/SET HMTs containing the MLL1 isoform and the growth of leukemia cells bearing rearrangements of the gene encoding MLL1. These findings led Vanderbilt Basic Sciences investigators William Tansey, Stephen Fesik, and Scott Hiebert and their labs to search for highly potent WIN site inhibitors and explore their effects on cancer cells. They began by screening a 13,800-member small molecule library in an NMR-based assay for compounds that inhibit binding of WDR5 to the MLL1 WIN motif peptide. They chose two molecules from the 47 hits identified by the screen for further structure-based design, ultimately discovering C3 and C6, which bound to WDR5 with high affinity (Kd= 1.3 and 0.1 nM, respectively). Using these probe molecules, they showed that WIN site inhibition suppressed the growth of many cell lines, particularly those containing rearrangements of the gene encoding MLL1 and those expressing wild-type p53. In most sensitive cell lines, exposure to the compounds resulted in death by apoptosis; however, suppression of p53 expression conveyed resistance to these toxic effects. Using RNA-seq, the researchers found that C6 exposure led to increased and decreased expression of 72 and 462 genes, respectively. Suppressed genes were primarily associated with protein synthesis, DNA replication, and cell cycle regulation. Chromatin immunoprecipitation coupled with next-generation sequencing confirmed binding of WDR5 in the vicinity of genes highly associated with ribosomal protein biosynthsis, and C6 suppressed the expression of these genes. Consistently, the investigators showed that WIN site inhibition disrupted protein synthesis, leading to nucleolar stress, induction of TP53gene expression, and increased levels of p53 protein. Precision nuclear run-on sequencing revealed that WIN site inhibition led to a reduction in active RNA polymerases associated with 47 transcription units, almost all of which were also sites of WDR5 binding and 70% of which coded for ribosomal subunit proteins. Interestingly, no effect on H3K4 methylation resulted from WIN site inhibition despite the fact that both C3 and C6 blocked the activity of MLL1-containing HMTs. Together, the findings, suggest that the primary function of the WIN site is to facilitate binding of WDR5 to chromatin and that disruption of that binding leads to reduction in gene transcription, particularly for genes encoding ribosomal proteins. The result is failure of protein synthesis, nucleolar stress, induction of TP53, and ultimately p53-mediated apoptosis. This valuable information lays the groundwork for the complete evaluation of the WIN site as a target for cancer chemotherapy. The work is published in the journal Cell Reports[E. R. Aho, et al. (2019) Cell Reports, 26, 2916].