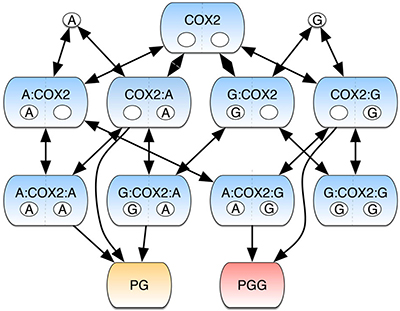
Multi-input/multi-output (MIMO) signaling systems, defined as networks that coordinate numerous inputs (e.g., enzyme substrates, receptor ligands, allosteric modulators) to yield multiple outputs (e.g., products, secondary signaling molecules, etc.) are common in living cells and critical for the modulation of all cellular processes. Understanding the dynamics of these complicated systems has proven incredibly difficult, as they have defied numerous attempts to explain complex and often juxtaposed outcomes. This led Basic Sciences investigators Carlos Lopez and Larry Marnett, their collaborator Eric Deeds (University of Kansas), and their labs to use the allosteric enzyme cyclooxygenase-2 (COX-2) as an interesting but tractable model system with which to computationally explore information transfer within a MIMO system.
COX-2 converts the polyunsaturated fatty acid arachidonic acid (AA) and its glyceryl ester (2-AG) into prostaglandins (PGs) and prostaglandin glyceryl esters (PGGs), respectively. Both PGs and PGGs are lipid signaling molecules possessing a wide range of biological activities. In addition, 2-AG plays an important role as a ligand in the cannabinoid signaling system, and COX-2-dependent metabolism of 2-AG inactivates this biological activity. Thus, COX-2 works at the crossroads of multiple pathways in a complex metabolic network that can direct the cell toward a range of alternate outcomes. To address how COX-2 integrates information from two inputs (AA and 2-AG), to yield two outputs (PGs and PGGs), the Lopez lab constructed the COX-2 Reaction Model (CORM), which described all possible reactions and complexes that could be formed by binding of AA or 2-AG to the catalytic and/or allosteric site of the enzyme. They then calibrated the model by applying a Bayesian parameter inference approach to kinetic data generated in the Marnett lab that measured the formation of PGs and PGGs from combinations of AA and 2-AG present over a wide range of concentrations.
The results suggested that binding of AA or 2-AG was more highly favored to occur first at the catalytic than at the allosteric site. Binding of either substrate at the allosteric site promoted conversion of AA to PGs while suppressing conversion of 2-AG to PGGs at the catalytic site. The Lopez lab then used CORM to evaluate the flow of information through the COX-2 reaction network. They discovered that of 6 possible pathways for PG formation, the enzyme primarily used only 4, and that of 4 possible pathways for PGG formation, the enzyme used only 2. The “pathway entropy”, a measure of the number of possible pathways utilized at any one time, depended on the concentrations of the inputs. Similarly, the system’s “mutual information”, which quantifies how knowledge of a system output provides information about a system input was highly dependent on the absolute and relative concentrations of AA and 2-AG as well as the degree to which the two inputs were correlated with each other. The findings confirmed that COX-2 integrates the signals conveyed by the presence of AA and 2-AG to yield outputs in the form of PG and PGG products. Information transmitted by the network depends both on the absolute and relative levels of the inputs as well as the correlation between the inputs. We look forward to further work applying these new concepts to PG and PGG biosynthesis in vivo. The work is published in the journal NPJ Systems Biology and Applications [E. M. Shockley, et al. (2019) NPJ Syst. Biol. Appl.5, 23;