Research
In human cells, ~120,000 replication forks are assembled during each cell cycle and traverse the genome. Collectively, these macromolecular machines completely synthesize the genome Failure to replicate even a ~10 base pair stretch of DNA can lead to catastrophic chromosome mis-segregation during the subsequent mitosis or activate error-prone DNA repair pathways that would elevate the mutation rate. It is thus crucial that every last stretch of DNA is replicated.
Complete synthesis of the genome poses two unique problems. First, mechanisms that ensure DNA synthesis at earlier stages of DNA replication can no longer once duplication of the genome approaches completion. Second, short stretches of unreplicated DNA cannot be detected by human cells. Thus, the mechanisms involved in completion of DNA synthesis need to be exquisitely accurate and we are interested in understanding how these events occur.
Termination of DNA replication
‘Termination’ of DNA replication occurs when two replication forks converge head-on upon the same stretch of DNA to complete DNA synthesis. This process occurs ~60,000 times per cell cycle and ensures that DNA synthesis is completed. Termination in bacteria and viruses can lead to under- or over-replication of DNA. In contrast, termination in vertebrates does not typically involve these events, suggesting that vertebrates possess specific mechanisms to ensure accurate termination.
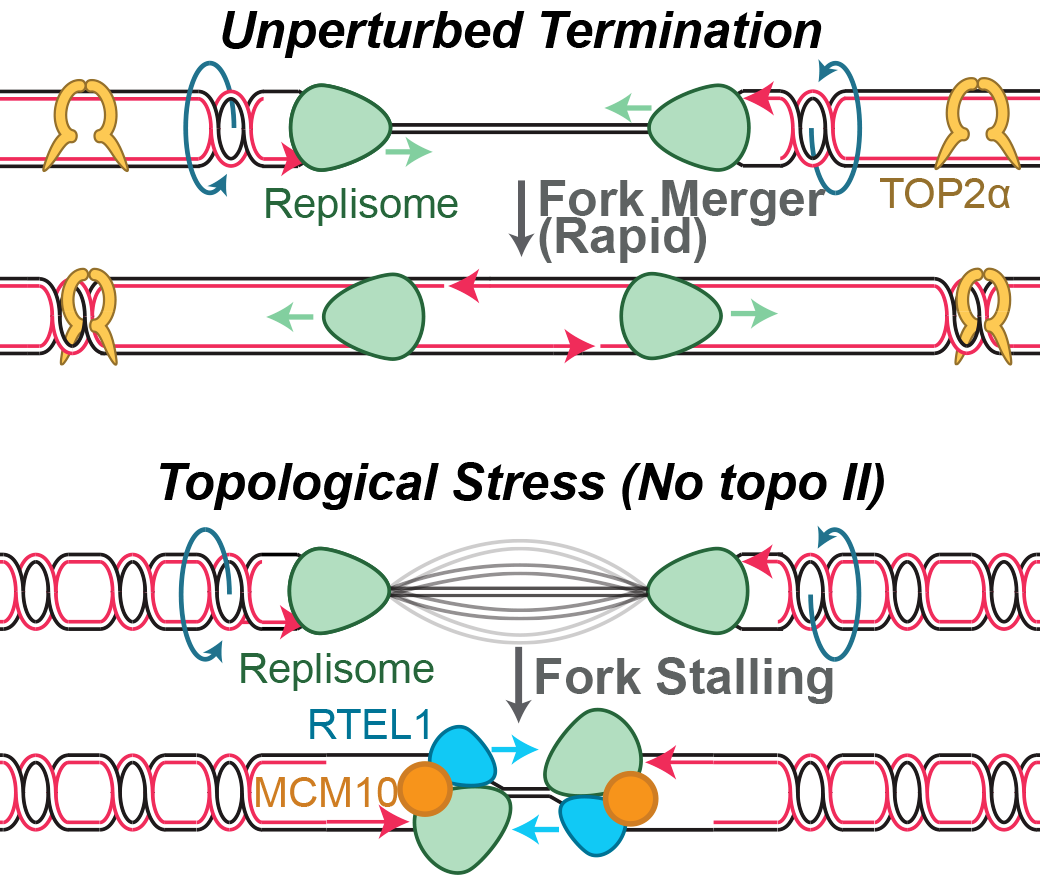
We are interested in trying to understand how vertebrate termination occurs without stalling or slowing of DNA synthesis. We found that topoisomerases play and important role in facilitating termination by preventing build-up of topological stress, but when topoisomerases fail replication then forks stall during termination. When this occurs, a back-up pathway involving stalled fork response proteins is activated to ensure replication fork stalling is overcome so that termination can be completed. Thus, our research has identified multiple pathways that ensure accurate termination, and we are investigating these pathways in more detail.
Health relevance: Topoisomerases are crucial for termination and are also a major target for cancer chemotherapy. We are interested in how topoisomerase usage is determined and how cancer chemotherapeutics that target topoisomerases work. More broadly, understanding termination will provide a better picture of cancer development because most mutations in cancer cells arise from defects in DNA replication.
Replication Restart
Endogenous and exogenous genotoxins can cause up to ~10,000 replication forks to stall during each S phase. In response, the replication fork junction is regressed and nascent DNA strands are extruded (‘fork reversal’). Nascent DNA strands are also subject to degradation by nucleases (‘nascent strand degradation’). Fork reversal and nascent strand degradation are thought to allow replication forks to overcome DNA lesions and restart DNA synthesis, in order to promote genome stability. However, fork reversal and nascent strand degradation can also cause genome instability when they occur excessively. It is thus important to understand how these processes are triggered efficiently, but not spuriously. Additionally, fork reversal and nascent strand degradation involve many proteins that are essential for viability and/or implicated in disease. Understanding these processes is therefore crucial to understanding how these important proteins function.
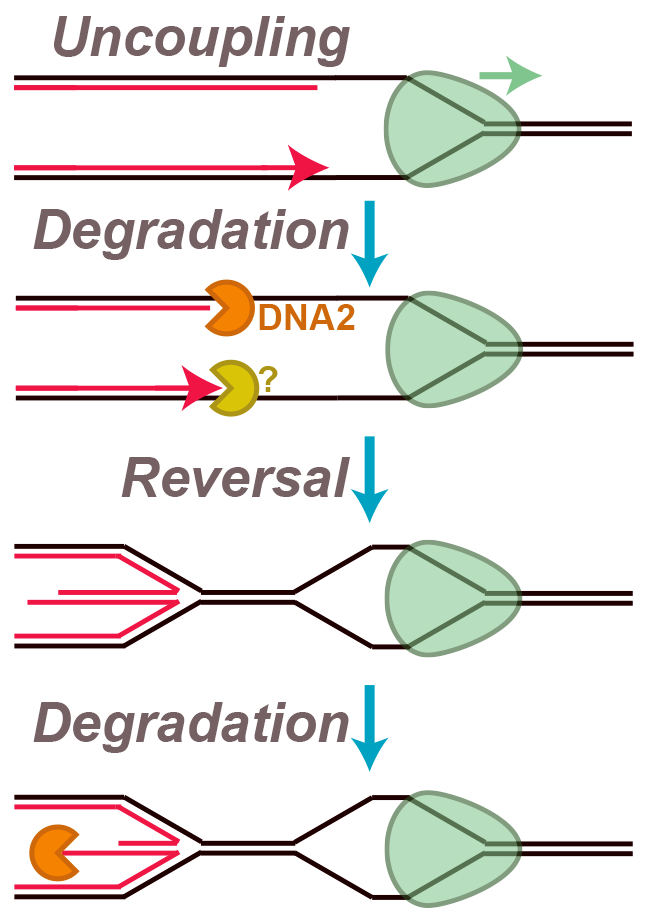
We are interested in trying to understand how fork reversal and nascent strand degradation are triggered and how exactly the underlying molecular events unfold. We recently developed a new in vitro approach to study nascent strand degradation and fork reversal, which we corroborated with cellular experiments. Surprisingly, we found that a subset of events involved in fork stalling are sufficient to cause fork reversal and nascent strand degradation. Specifically, we showed that these events can be triggered by replication fork ‘uncoupling’, which occurs when the DNA polymerases stall but the replication fork helicase keeps unwinding. We also identified an additional degradation step, prior to fork reversal, and found that the replicative helicase is retained throughout this process. We are now studying exactly how uncoupling triggers these events, and how different proteins participate in fork reversal and nascent strand degradation.
Health relevance: Fork reversal and nascent strand degradation are a primary response to genotoxins derived from either the environment (e.g. UV and ionizing radiation) or natural cellular metabolism (e.g. reactive oxygen species). It is important to understand how these processes are triggered and how they work so that we can better evaluate the risk of exposure to genotoxins. Fork reversal and nascent strand degradation involve tumor suppressors (e.g. BRCA1, BRCA2). Studying the role of these proteins during fork reversal and nascent strand degradation will allow us to better understand how they mitigate cancer development. Finally, defects in replication restart are targeted during cancer chemotherapy so understanding these pathways should inform the development of improved cancer chemotherapies.
Telomere Replication
At each end of every chromosome is a ‘telomere’, which is a nucleoprotein cap that distinguishes chromosome ends from DNA double strand breaks. A multi-subunit protein complex called ‘Shelterin’ binds to telomeric repeat sequences and ensures that telomeres do not trigger checkpoint activation or undergo the processing associated with double-strand break repair. Because of the unique structure of telomeres and their position at the chromosome end, it is impossible for the replication machinery to fully replicate telomeres. Accordingly, telomeres shorten with every genome duplication event until they become so short that they no longer function. Cells then recognize chromosome ends as DNA breaks, which prevents the cell from dividing further. This process is unavoidable and contributes to the aging process in multicellular organisms.
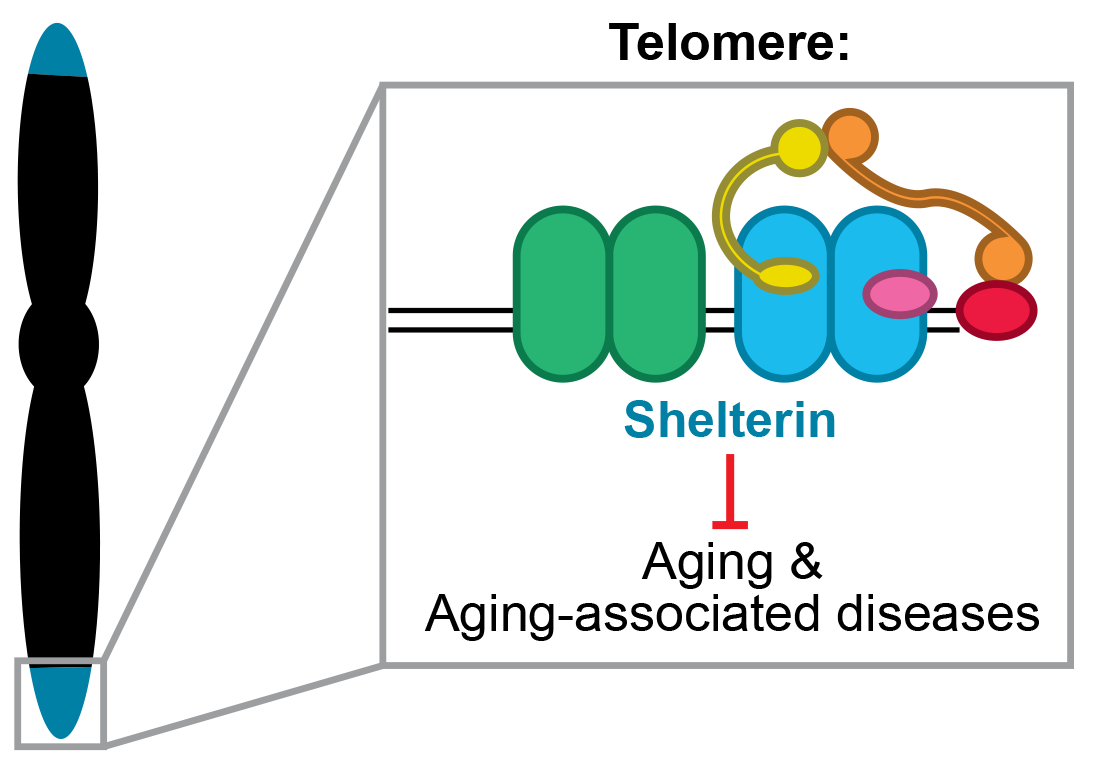
Telomere replication is the one clearly defined situation where completion of DNA synthesis is not completely accurate and results in unavoidable sequence loss. Our goal is to understand how sequence loss is minimized during telomere replication in order to maximize the fidelity of telomere replication. We are also interested in exactly which features of short telomeres render them non-functional.
Health relevance: Telomere shortening plays a causal role in aging- and aging-related diseases. Understanding how the rate of telomere shortening is controlled and the minimum requirements for telomere function will allow to develop approaches to better-evaluate individuals’ health status.