Research Focus
Nakagawa lab research program
Introduction:
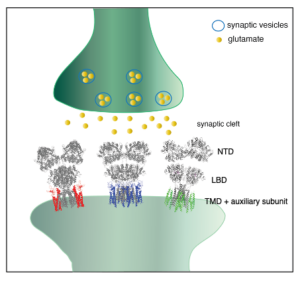
Synaptic transmission mediates information transfer between neurons in the brain. When action potentials reach the presynaptic terminal, the neurotransmitter stored in the synaptic vesicles is released to the synaptic cleft, which in turn activates its receptors in the postsynaptic membrane. The amino acid L-glutamate is the major excitatory neurotransmitter in mammalian brain synapses. The ligand-gated ion channels activated by L-glutamate are the members of ionotropic glutamate receptors (iGluRs), which consists of NMDA, AMPA, and kainate receptors. The activation of the iGluRs by L-glutamate induces the ion channel to open briefly, generating a pulse of depolarization in the postsynaptic membrane.
The amplitudes of synaptic response, as well as the time course of activation and decay varies substantially under different circumstances. It is widely recognized that reversible tuning (or modulation) of the synaptic response, known as synaptic plasticity, is a critical cellular mechanism underlying learning and memory in neuronal circuits. We investigate the molecular mechanism underlying synaptic transmission and plasticity mediated by the iGluRs.
In humans, dysfunction of iGluRs is related to a variety of neurological and psychiatric disorders, including schizophrenia, Alzheimer’s disease, ALS, intellectual disability, brain tumor, seizure, limbic encephalitis, CNS lupus, and Rasmussen’s encephalitis. Understanding the precise molecular basis for iGluR function is expected to facilitate identification of new therapeutic targets and compounds that could control iGluR functions in disease conditions.
We focus to provide structural and biophysical basis for the signaling mediated by the iGluRs. Most of our current research is on the AMPA-subtype of iGluRs (AMPA receptors, or AMPARs), which mediates most of the excitatory synaptic transmission in mammalian brain (Figure 1).
Initial structure of native AMPAR complexes:
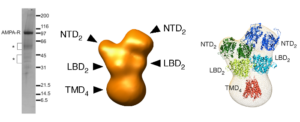
In 2005, many years before the cryo-EM resolution revolution, we determined the structure of the AMPAR complexes purified from brain at the resolution of 20-30Å (Nakagawa et al., Nature, 2005 and Nakagawa et al., Biological Chemistry, 2006). The dimer-of-dimer global architecture of GluA2 containing, presumably heterotetrameric, native AMPARs was demonstrated (Figure 2). Conformational changes related to desensitization was shown; the two N-terminal domain (NTD) dimers undergo lateral displacement after prolonged exposure to the agonist L-glutamate. The study also revealed the presence of auxiliary subunits (TARPs) in the native AMPARs. The work provides the first glimpse of the ionotropic glutamate receptor architecture (see Mayer Nat Struct Mol Biol. 2005 Mar;12(3):208-9 and Hansen et al., 2021) and forms the foundation of our labs research programs.
Research Projects:
(1) Structure and functional mechanism of AMPAR auxiliary subunit cornichon.
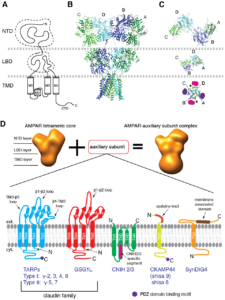
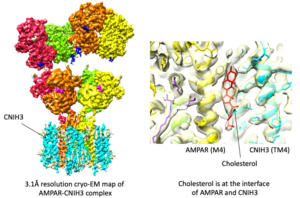
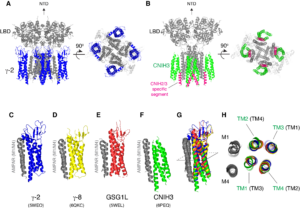
Signals transduced by AMPAR-auxiliary subunit complexes are critical for synaptic plasticity, learning, and memory. The two major categories of AMPAR auxiliary subunits are transmembrane AMPAR regulatory proteins (TARPs) and cornichon homologs (CNIHs); these subunits share little homology and play distinct roles in controlling ion channel gating and trafficking of AMPAR (Figure 3). In 2014, we defined sub-regions in CNIH3 auxiliary subunit and AMPAR required for binding and gating modulation, and reported methods to prepare recombinant AMPAR-auxiliary subunit complexes for structural studies (Shanks et al., J. Neurosci., 2014). In 2017, we conducted a reciprocal mutational analysis, identifying key residues of AMPAR that are critical for the action of auxiliary subunits CNIH3 and TARPg2 (Hawken et al., J. Physiol., 2017). In 2019 my lab determined high-resolution cryo-EM structures of the AMPAR in complex with CNIH3 (Nakagawa., Science, 2019: A single author paper in Science) (Figure 4, left). Contrary to its predicted membrane topology, CNIH3 lacks an extracellular domain and instead contains four membrane-spanning helices (Figure 5). The protein-protein interaction interface that dictates channel modulation and the lipids surrounding the complex are revealed (Figure 4, right). These results provide paradigm-shifting insights into the molecular mechanism for ion channel modulation and assembly of AMPAR/CNIH3 complexes. Moreover, it is the first molecular structure of the cornichon family of membrane proteins, conserved from yeasts to mammals and critical for certain vesicle trafficking and secretion (see commentary by Schwenk and Fakler, Science, 2019). We continue to study the mechanism of cornichon family in AMPAR gating modulation and biogenesis.
(2) In vivo function of AMPAR auxiliary subunit
Various auxiliary subunits amplify AMPAR function by regulating ion channel gating and/or trafficking. In 2012, we identified a new AMPAR auxiliary factor GSG1L (Shanks, Savas, and Maruo et al., Cell Reports, 2012) (Figures 6 and 7).
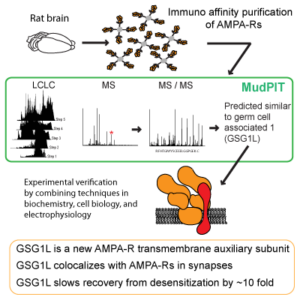
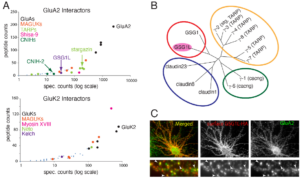
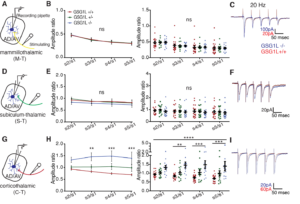
GSG1L uniquely delays the AMPAR recovery from desensitization, at a magnitude greater than any of the known auxiliary subunits, thus functioning as negative regulator of AMPARs. To understand the in vivo function of GSG1L we generated a GSG1L KO mice line and studied the phenotypes (Kamalova et al., Cell Reports, 2020). In the GSG1L KO mice, short-term plasticity was abnormally regulated in the anterior thalamus, where GSG1L expression is high in wild type (Figure 8). Consistent with the negative regulatory role of GSG1L, the GSG1L KO mice are susceptible to seizures. We continue to study the function of GSG1L in synapses, circuit activity, and behavior. The most recent collaborative work on GSG1L biology is (Perozzo et al., Nat. Commun., 2023).
(3) Subunit assembly mechanism of AMPA receptors
In 2010, we determined the single particle EM structures of the dimeric subunit assembly intermediates of the AMPA-Rs (Shanks et al., J. Neurosci., 2010). Our results suggest that specific global domain arrangement of the dimer intermediate is required for the efficient tetramerization. Furthermore, we proposed a global domain arrangement in the tetrameric AMPA-R, which requires the domain swap observed in the mature tetrameric AMPA receptors (Sobolevsky et al., Nature, 2009). A set of membrane associated proteins (FRRS1l, CPT1c, and ABHD6) preferentially associates the AMPA receptors in the biogenesis pathway, reported by the Fakler group (Schwenk et al., Neuron, 2019). We plan to take structural approach to elucidate the AMPA receptor biogenesis pathway to further understand the mechanism governing AMPA receptor co-assembly with auxiliary subunits.
(4) Mechanism of ion permeation in iGluRs.
Transporting ions across the membrane in response to activation by neurotransmitter is the fundamental function of most iGluRs. In AMPARs, their auxiliary subunits bidirectionally modulate ion channel conductance, which is directly related to changes in how ion travels through the channel pore. Accurate information of the pore geometry and distribution of water and ions in the pore is expected to advance our understanding of the mechanisms underlying modulation of channel function and facilitate development of new therapeutics targeting the pore. Indeed, the pore contains binding sites for compounds that block channel function. Despite the importance, the ion permeation mechanism of iGluRs remain elusive because information about ion biding sites in the pore is missing. To tackle this question, we have investigated the distribution of non-protein densities in the open pore by cryo-EM and how they translate to modulating ion permeation.
(5) Drug discovery targeting AMPAR function.
The progresses made in the structural biology and ion channel physiology of iGluRs opened an unprecedented opportunity to explore the mechanism of compounds of therapeutic relevance. In 2017, we reported our results on the high throughput drug screening aimed to identify new compounds that are specific to auxiliary subunits of AMPARs (Azumaya et al., PlosONE, 2017). We plan to solve the structures of the AMPAR/auxiliary subunit complex in the presence of the identified compounds. The outcomes of these efforts are expected to identify new binding pockets for drugs and facilitate structure-based improvements of the lead compounds.
(6) Other areas of interests
(a) function of lipids in ion channels, (b) time resolved cryo-EM of ion channels, (c) molecular dynamic simulation of ion channels, (d) cryo-electron tomography of synaptic macromolecular complexes, and (e) collaborative work related to neuroscience using cryo-EM.
Our Laboratory and the Cryo-EM facility
Terunaga Nakagawa is currently the Scientific Director of Cryo-EM at Center for Structural Biology (CSB) at Vanderbilt University. He collaborated extensively with the Dean’s Office, CSB, and various colleagues to establish the current cryo-EM facility that houses a Titan Krios G4, and a Glacios cryo-TEM (obtained by NIH S10 OD030292, PI:Nakagawa) among other associated instruments for specimen preparation. Thanks to the team efforts made by the Facility Co-Directors Melissa Chambers and Scott Collier, and Staff Scientist Mariam Haider, a substantial progress was made during the past 10 years at Vanderbilt. Currently, many laboratories are using the cryo-EM facility at Vanderbilt and producing exciting results (see link to CSB cryo-EM facility publication page).