Research
Research Focus
The overarching goal of the research in our lab is to understand the gene regulatory mechanisms governing erythropoietin (Epo)-induced terminal erythroid differentiation.
Description
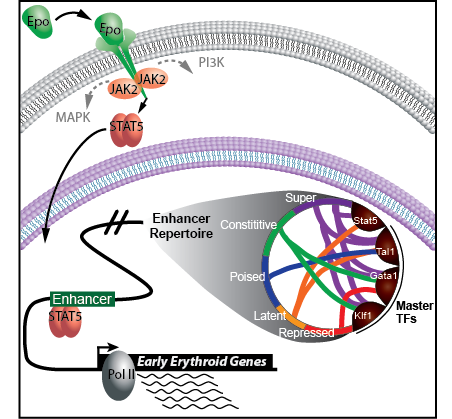
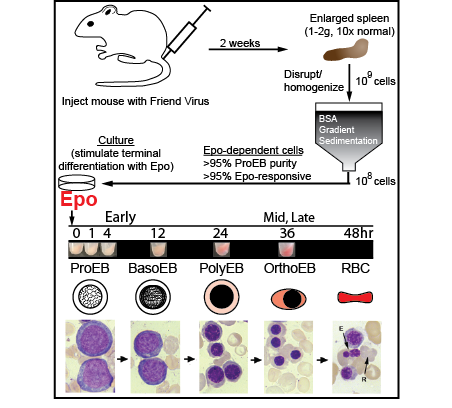
Erythropoietin (Epo) is the primary hormone regulator that controls erythroid cell maturation, a process that is required for the daily replenishment of nearly 1% (200 billion) of the circulating red blood cells. Despite the widespread clinical use of Epo to treat chronic anemia, we still do not fundamentally understand how Epo works to promote terminal erythroid differentiation. A central goal of our research is to understand how Epo signaling controls transcriptional mechanisms governing terminal erythroid differentiation. The lab’s research involves (1) development of cutting edge functional genomic and computational tools, (2) application of these tools to dissect fundamental erythroid cell biology, and (3) translation of key discoveries into new therapeutics and treatment strategies. Research is especially focused on targeting transcriptional regulators in primary murine erythroid precursors and tracking them over time in response to Epo. The laboratory spans the gap between wet and dry lab disciplines in that we first generate high-quality sequencing libraries and also have the expertise to analyze our data using custom computational pipelines.
Current Projects
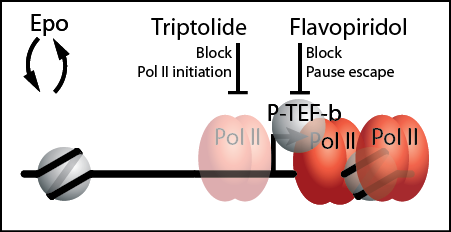
1. Determine how Epo signaling controls RNA polymerase II pausing and elongation.
Transcription elongation has emerged as a major theme in regulating some key erythroid genes during erythropoiesis, but whether the cohort of Epo-induced erythroid genes are rate-limited at Pol II recruitment or pause release has not been examined. Based on our preliminary data, we hypothesize that Epo signaling controls the rate-limiting steps Pol II initiation and pause release at Epo-induced genes. To test this hypothesis, we will employ a pharmacological “push/pull” strategy detailed below to further characterize the underlying mechanisms for how Epo controls transcriptional elongation.
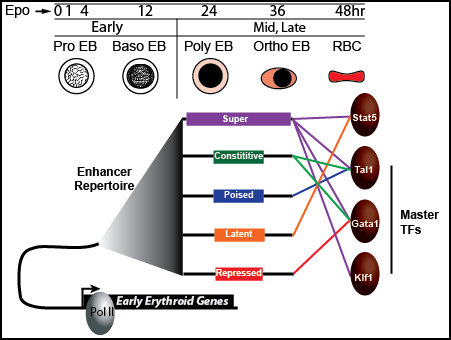
2. Elucidate how early Epo-responsive genes are regulated by transcription factors and enhancers.
We have identified ~60,000 putative enhancer regions in erythroid cells prior to Epo stimulation using hidden Markov modeling of epigenetic patterns. Which transcription factors bind these enhancers and which of these enhancers are Epo-responsive remains unclear, and thus we have designed experiments are designed to address this. Finally, we would like to investigate the role TFs play in altering the enhancer repertoire in response to Epo.