By Alexandra Fuller
You might break chicken eggs for your breakfast, but the lab of James Dewar (Biochemistry) breaks frog eggs to better understand the mechanisms cells use to terminate DNA replication. Published in Cell Reports, the Dewar lab found that topoisomerase IIα paves the way for DNA replication termination in vertebrates by keeping DNA tangle free. Darren Heintzman and Lillian Campos-Doerfler are co-first authors of this work.
DNA’s twisted structure makes it tangle easily; DNA is stored in a tightly wound, compact form so that it fits within the cell. Topoisomerases help cells keep their genomes in top shape by transiently cutting the DNA, removing any knots and tangles in it, and putting it back together again, leaving it as good as new. Topoisomerase IIα is the primary enzyme active during DNA replication.
Compromising between storage and access presents challenges: to make new cells, DNA must be opened up and replicated to provide each daughter cell with its own copy of DNA; opening a segment of DNA, however, twists other areas, creates tension, and may cause tangles. Unresolved DNA tangles can lead to many problems, including the possibility of molecular “hangovers,” as described by Dewar. Much like with hangovers, the accumulation of molecular problems (DNA tangles) causes undesirable effects later, during termination.
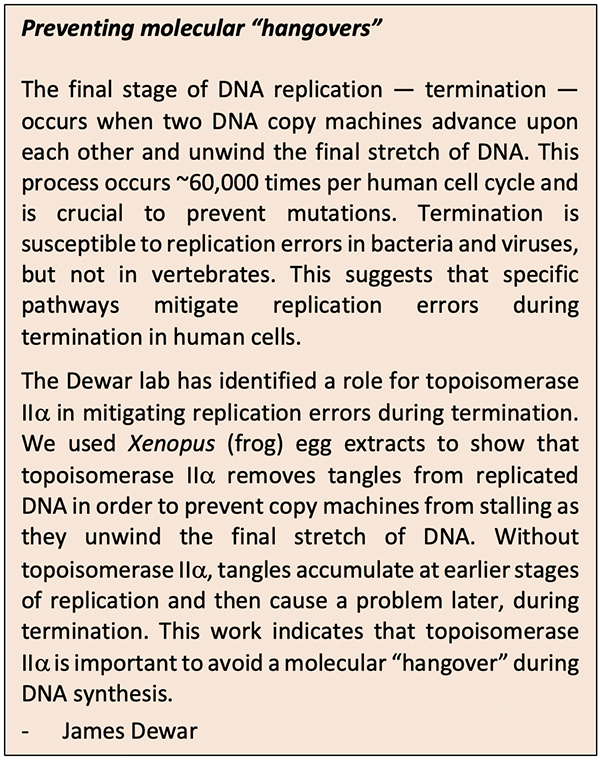 The Dewar lab studies the molecular events that allow for successful replication termination in vertebrates. Termination is poorly understood compared to the earlier stages of DNA replication, and most of the work in this field has been performed in bacteria, viruses, and yeast rather than vertebrates.
The Dewar lab studies the molecular events that allow for successful replication termination in vertebrates. Termination is poorly understood compared to the earlier stages of DNA replication, and most of the work in this field has been performed in bacteria, viruses, and yeast rather than vertebrates.
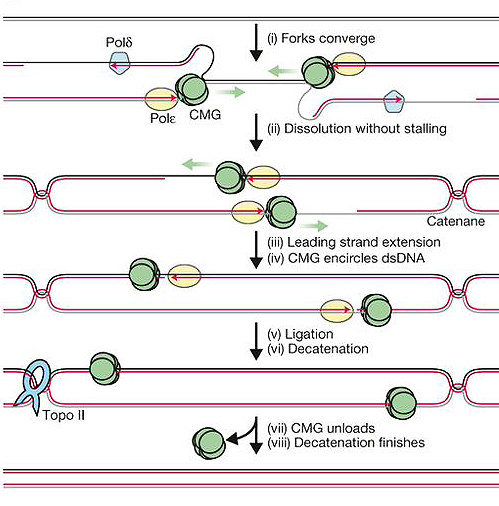
To determine topoisomerase IIα’s part in vertebrate replication termination, the Dewar lab used an ingenious frog egg-based system. Termination was broken down into separate events that included copying the final stretch of DNA and resolving tangles.
Vertebrate copy machinery (the proteins and enzymes used to copy DNA) provided by frog egg extracts was used to replicate DNA plasmids (circular bacterial DNA). A reversible barrier was built into these plasmids so that replication could be “paused” before the final stretch of DNA. Having control over when to “unpause” the replication process allowed the researchers to ask how long it took to synthesize the final stretch of DNA if topoisomerase IIα were removed or inhibited.
Topoisomerase IIα was either depleted throughout DNA replication or inhibited just prior to termination; eliminating the enzyme throughout replication caused longer delays during termination compared to inhibiting it just prior to termination. These findings suggest that removing or inhibiting topoisomerase IIα during earlier stages of DNA replication impairs and delays termination later on, but does not prevent its completion. Thus, the researchers concluded that the last bit of DNA can be replicated independently of topoisomerase IIα.
Topoisomerase IIα cuts both strands of DNA, allowing them to separate and untangle. In agreement with this known function, DNA tangles were not resolved (untangled) when topoisomerase IIα was removed, as tangles were observed even when the enzyme’s activity was blocked just prior to termination. When the researchers added back topoisomerase IIα to extracts depleted of the enzyme, the tangles were resolved and DNA copying during termination occurred at a normal pace. Delays in termination were still observed if topoisomerase IIα with an impaired DNA-cutting ability was added. These results suggest that DNA restructuring by topoisomerase IIα may allow termination to occur smoothly.
Frog and human DNA-copying machinery are similar enough that these results may apply to us as well; however, the bacterial DNA used in these experiments is packaged differently than vertebrate DNA, and further work is needed to show that topoisomerase IIα behaves similarly in frogs and humans.
The combined findings presented in this paper support a model in which topoisomerase IIα untangles DNA throughout replication, allowing for timely DNA replication termination. Without topoisomerase IIα, unresolved DNA tangles build up and delay replication termination. Understanding termination is important due to the severe consequences that can occur if replication does not go exactly according to plan, as this can cause DNA damage and cell death. Therapeutics that “poison” topoisomerase IIα intentionally cause DNA damage to kill rapidly reproducing cancer cells but also may cause heart complications and secondary cancers; acquiring an increased understanding of how DNA replication happens could lead to the development of better therapies.
Funding from the National Institutes of Health.
With contributions from Francisco Guillermo Rodriguez.