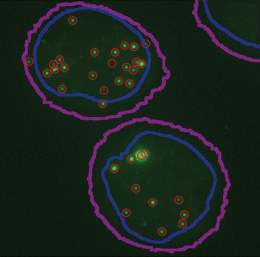
Modern biology allows scientists to fluorescently tag specific and individual RNA molecules or proteins, and the resulting fluorescence can be quantified under a microscope, serving as a proxy of how much of that RNA or protein is present in a given location. The resulting visualizations generate large data sets that need to be quantified to extract meaningful biological insights. According to a recent paper published in Genome Biology, however, the “manual or semi-automated quantification of such images is labor intensive, biased, and difficult to reproduce,” not to mention that it often limits the amount of interpretable information that can be gleaned from the sample.

To overcome limitations of current image analysis methods, the lab of Gregor Neuert, associate professor of molecular physiology and biophysics and senior author of the recent study, developed TrueSpot, a software tool that automates the detection and quantification of fluorescent spots. Although TrueSpot was designed for high-throughput processing of images using a computer cluster (a connected set of computers that pool their processing power to help with solving complex problems), it has a simple graphical interface that users can access through a desktop and can handle lighter loads.
Given that fluorescence microscopy is used across an enormous range of biological, biomedical, and physical research, TrueSpot has the capacity to overhaul the quantity and quality of usable data that can be obtained from it.
We connected with Neuert, who shared more details about TrueSpot and his lab’s latest paper.
What issue does your research address?
We address the challenge of how to accurately quantify signal puncta (discrete spots) in fluorescent imaging, particularly in RNA fluorescent in situ hybridization (known as RNA-FISH), immunofluorescence, and phase condensate experiments. Existing tools often require manual adjustments and are inconsistent across different imaging parameters, which leads to labor-intensive processes and variability in results.

What was unique about your approach to the research?
Blythe Hospelhorn, a graduate student in my lab, developed TrueSpot (with initial work from Ben Kesler and data from Hossein Jashnsaz). TrueSpot is a fully automated software tool designed for the robust detection and quantification of signal puncta in both 2D and 3D images without the need for manual intervention. This automation aims to reduce subjectivity and improve consistency in results. Vanderbilt’s ACCRE compute cluster was instrumental in analyzing the data.
What were your top three findings?
- TrueSpot outperformed other existing tools like Big-FISH and RS-FISH in both simulated and experimental datasets, showing higher precision and recall. Precision and recall (sensitivity) are used as benchmarking measures to determine the ideal parameters for analyzing each data set.
- Setting a threshold at which algorithms consider puncta to be true signals versus just noise in an image has been a challenging aspect of image analysis. We designed TrueSpot to use an automated threshold selection algorithm that uses multiple approaches to set a signal threshold for individual data sets. TrueSpot outperformed other automated tools particularly on images with varying background noise and signal intensity.
- Microscopy can generate an image of a cross-section of a cell or tissue of interest, but if you “stack” those images together, you can get a good idea of what the whole cell or tissue looks like in three dimensions. Currently, many existing deep learning–based tools only operate on 2D images instead of on 3D image stacks. While TrueSpot can accept either a 2D image or a 3D image stack, we found that 3D image processing with TrueSpot yields more accurate detection of RNA signals than relying on the traditional analysis derived from looking at 2D images.
Who or what made the difference in your research?
The support from interdisciplinary collaborators and feedback from peers made an invaluable difference. The collaborative environment at Vanderbilt fostered innovation and iterative testing and led to things like insights from manual thresholding experiences.
What do you hope will be achieved with the research results on the short term?
We aim to provide researchers with an efficient, user-friendly tool that enhances the accuracy and throughput of RNA quantification in single-cell studies, which will facilitate larger and more reproducible datasets.
What are your highest translational/clinical aspirations that might come from this research?
We hope that TrueSpot will advance the field of molecular biology and molecular pathology by enabling clearer insights into gene expression dynamics and disease mechanisms. The improved RNA visualization that can be achieved with TrueSpot will hopefully ultimately contribute to improved personalized medicine strategies.
Where is this research taking you next?
This research is leading toward further enhancements of TrueSpot, including machine learning integrations to improve its performance on diverse datasets. I personally look forward to refining the software and collaborating with fellow researchers to apply it to new biological questions.
Go deeper
The paper “TrueSpot: a robust automated tool for quantifying signal puncta in fluorescent imaging” was published in Genome Biology in September 2025.
All MATLAB code comprising TrueSpot and the Neuert lab’s software pipeline is open source (under the MIT license) and available on Zenodo and GitHub.
Funding
This research used funds from the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of General Medical Sciences, the National Institutes of Health Office of the Director, and a Vanderbilt University School of Medicine Basic Sciences Dean’s Faculty Fellowship.
Open Access
The study was published open access through a transformative agreement negotiated by Vanderbilt University’s Jean and Alexander Heard Libraries. Transformative agreements eliminate traditional paywalls and remove the obstacle of article processing charges, ensuring immediate and unrestricted access to research worldwide. Vanderbilt authors can learn more about the Heard Libraries’ agreements supporting open access publishing in this research guide.