CLASPs Are Required for Proper Microtubule Localization of End-Binding Proteins
CLASPs Are Required for Proper Microtubule Localization of End-Binding Proteins
Ashley D. Grimaldi1, Takahisa Maki2, Benjamin P. Fitton4, Daniel Roth4, Dmitry Yampolsky1, Michael W. Davidson3, Tatyana Svitkina5, Anne Straube4, Ikuko Hayashi2, Irina Kaverina
Summary
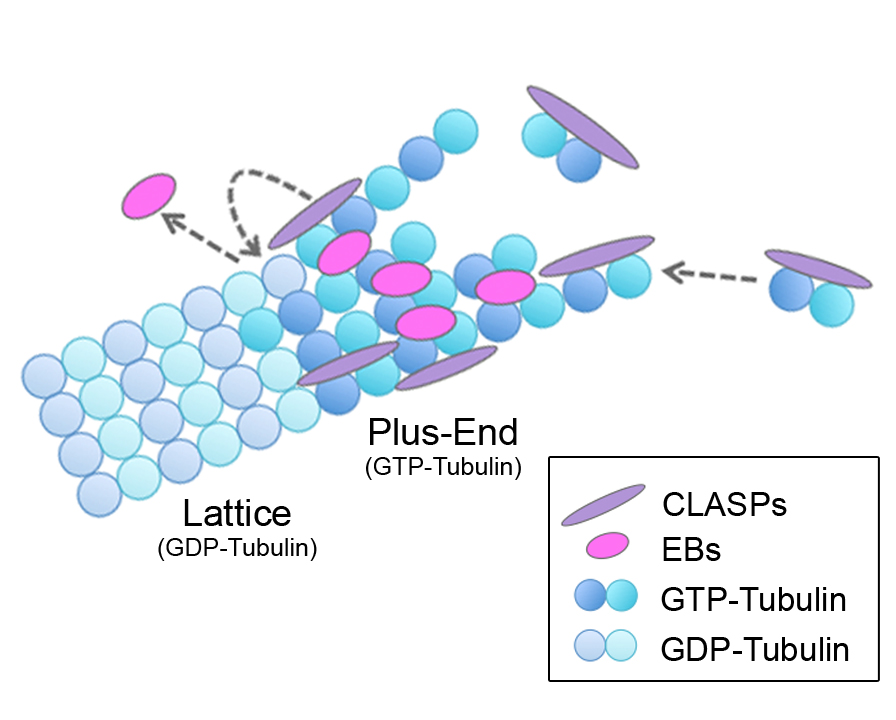
Microtubule (MT) plus-end tracking proteins (+TIPs) preferentially localize to MT plus ends. End-binding proteins (EBs) are master regulators of the +TIP complex; however, it is unknown whether EBs are regulated by other +TIPs. Here, we show that cytoplasmic linker-associated proteins (CLASPs) modulate EB localization at MTs. In CLASP-depleted cells, EBs localized along the MT lattice in addition to plus ends. The MT-binding region of CLASP was sufficient for restoring normal EB localization, whereas neither EB-CLASP interactions nor EB tail-binding proteins are involved. In vitro assays revealed that CLASP directly functions to remove EB from MTs. Importantly, this effect occurs specifically during MT polymerization, but not at preformed MTs. Increased GTP-tubulin content within MTs in CLASP-depleted cells suggests that CLASPs facilitate GTP hydrolysis to reduce EB lattice binding. Together, these findings suggest that CLASPs influence the MT lattice itself to regulate EB and determine exclusive plus-end localization of EBs in cells.
Introduction
Microtubules (MTs) are inherently polar structures polymerized from GTP-tubulin heterodimers, which incorporate at the growing MT plus end. Plus-end tracking proteins (+TIPs) are a diverse group of conserved MT-associated proteins (MAPs) that specifically localize to the dynamic tips of MTs (Schuyler and Pellman, 2001). +TIPs are ideally positioned to regulate MT dynamics and many important MT-based processes. Of the +TIPs, end-binding proteins (EBs) are notable in their ability to autonomously recognize a transient feature at plus ends with great specificity (Bieling et al., 2007). In mammalian cells, there are three EB family members: EB1, EB2, and EB3. EB1 and EB3 are well studied and are similar in their structure, localization, and behavior, whereas EB2 is more divergent (Buey et al., 2011 and Komarova et al., 2009). Recent work has revealed that EBs localize to MT tips by sensing the nucleotide-state, as well as the specific conformation, of tubulin at MT plus ends (Maurer et al., 2011, Maurer et al., 2012 and Zanic et al., 2009). The majority of +TIPs require binding to the EB tail region, via various localization signals (SxIP, Honnappa et al., 2009; or CAP-Gly, Weisbrich et al., 2007), to achieve their specific comet-like localization (Akhmanova and Steinmetz, 2008); for this reason, EBs are considered the master regulators of the MT +TIP network.
Within this network, CLASPs are a unique class of +TIPs that exhibit two specific MT localizations. CLASPs support MT growth by binding plus ends and promoting rescues (Akhmanova et al., 2001 and Mimori-Kiyosue et al., 2005). CLASPs also stabilize MT subsets by binding along the MT lattice, for example pioneer MTs at the leading edge (Wittmann and Waterman-Storer, 2005) and Golgi-derived MTs (Efimov et al., 2007 and Miller et al., 2009). In mammalian cells, there are two redundant CLASP family members, CLASP1 and CLASP2, which rely on EB interactions (via the basic/SxIP region) to localize to MT tips (Mimori-Kiyosue et al., 2005). In fission yeast, although CLASP associates with EB1, its stabilizing effect on MTs is independent of other +TIPs suggesting that CLASP directly regulates MTs (Bratman and Chang, 2007). Of note, CLASPs also independently bind tubulin heterodimers and associate with the MT lattice through their TOG (tumor-overexpressed gene) domains (Al-Bassam et al., 2007 and Al-Bassam et al., 2010). Here, we report evidence that CLASPs are crucial determinants of proper EB localization at MTs in cells.