HGEN Student Resources
Resources for HGEN students
HGEN students draw on a wide range of research, educational and support resources from both Vanderbilt University and Vanderbilt University Medical Center.
VANDERBILT GENETICS INSTITUTE (VGI)
The VGI was established in January, 2015 to broaden and enhance the intellectual environment in genetics and genomics at Vanderbilt. Genetic and genomic sciences are – and should be – represented across multiple departments and centers at Vanderbilt. The VGI is the intellectual home for all groups doing genetic and genomic research at Vanderbilt. It has recruited and continues to recruit a substantial number of new faculty with a primary focus in both quantitative and molecular genetics and genomic sciences. The institute has established partnerships with multiple departments to spearhead new initiatives in genetics and genomics, including the construction of substantial new research infrastructure in computational genomics. Dr. Nancy Cox, a quantitative human geneticist with a long-standing research program in developing and applying novel analytic methods to genetic and genomic data in order to identify and characterize the genetic component to common human diseases, was recruited to lead the VGI. In addition to recruiting more faculty in this rapidly evolving field to Vanderbilt, the VGI continues to make substantial additional investments in genome interrogation in BioVU, Vanderbilt’s unique biobanking resource (see below) and in establishing partnership relationships with external data sources such as the UK BioBank and All of Us. The Vanderbilt Genetics Institute has substantial computing resources, including a total of 272 cores on 17 nodes with 1.6 Tb of Ram with 242 Tb of storage. A full suite of state-of-the-art computer programs for genetic and genomic analysis (sequence analysis pipelines, association analysis, linkage analysis, family reconstruction software, family-based methods of analysis) are maintained as well as general statistical software such as MATLAB, SAS, Statistica, R, S+. Database engines include Oracle, MySQL, and Access. In addition, we are able to access ACCRE, as described below for all computing activity.
THE ADVANCED COMPUTING CENTER FOR RESEARCH AND EDUCATION (ACCRE)
The Vanderbilt University Advanced Computing Center for Research and Education (ACCRE) is a researcher driven ‘collaboratory’, operated by and for Vanderbilt faculty. ACCRE was initially funded by a seed grant of $8.3M from Vanderbilt, with the expectation that the center would become largely self-supporting via contributions from individual Vanderbilt researchers. Over 800 researchers from over 40 campus departments and 5 schools have used ACCRE in their research and education programs. The computer cluster at ACCRE currently consists of over 7,000 processor cores and is growing. Funding for all hardware has come from external grants or startup funds contributed by collaboratory faculty. In addition to hardware, ACCRE has a staff of 11 support personnel who maintain and operate the cluster and provide user education and outreach.
The ACCRE high-performance computing cluster has over 7,000 processor cores (providing roughly 550 teraflops of single-precision floating point performance) and is growing. Nodes each have 24 – 256 GB of memory. Compute nodes all run a 64-bit Linux OS and have a 250 GB – 1 TB hard drive and dual copper gigabit Ethernet ports. Twenty-six compute nodes are each equipped with 4X Nvidia Titan X GPU cards, which provides an additional 933 teraflops of single-precision floating point performance. These nodes are also interconnected with a low-latency 40/56 Gb/s RoCE network. All compute nodes are monitored via Nagios. Resource management, scheduling of jobs, and usage tracking are handled by an integrated scheduling system by SLURM. These utilities include an “advance reservation” system that allows a block of nodes to be reserved for pre-specified periods of time (e.g., a class or lab session) for educational or research purposes.
IBM’s General Parallel File System (GPFS) is used for user home and data directories and scratch space. The ACCRE filesystem provides over 1 PB of usable disk space and can sustain more than 130 Gb/s of I/O bandwidth to the cluster. The home directories of all users are backed up daily to tape. The disk arrays are attached to a SAN fabric along with the storage nodes that then exports the file system to the rest of the cluster using a fully redundant design with no single point of failure.
The daily operation and maintenance of ACCRE is provided by eleven support personnel, including eight system administrators, programmers, and researchers with a combination of more than 60 years of computing experience. Support for system services are provided on a 24-hour basis for urgent issues, with on-call pager-based support on nights and weekends. Cluster uptime has been better than 95% over the past five years. An online support ticket system is used to track and resolve problems and user questions. ACCRE staff are responsible for maintaining core system hardware and software, networking, user support, tape backup services, disk storage, logistical storage development work, education, and management/finance support.
 ACCRE implements numerous security measures in order to maintain the privacy of user data. For example, firewalls filter cluster access from external hosts, passwords authenticate user login, and file permissions control data access on the user and group levels. File encryption software is available on the cluster and may be applied by users on a file-by-file basis for added security.
ACCRE implements numerous security measures in order to maintain the privacy of user data. For example, firewalls filter cluster access from external hosts, passwords authenticate user login, and file permissions control data access on the user and group levels. File encryption software is available on the cluster and may be applied by users on a file-by-file basis for added security.
Additional security features are enabled for data falling under International Traffic in Arms Regulation (ITAR), Export Administration Regulations (EAR), Protected Health Information (PHI), Research-related Health Information (RHI), or proprietary control. The Principal Investigator (PI) of each group completes a Disclosure for his/her research group specifying whether the group will work with ITAR, EAR, PHI, RHI, or proprietary data; new group users require explicit approval from the PI. This policy ensures that the PI controls user access to any restricted files and can educate users on the group’s internal security precautions. Groups working with ITAR, EAR, PHI, RHI, or proprietary data are noted with a special designation to differentiate them to cluster administrators, and permissions on these data types are limited to group readable and writeable only. If users inadvertently leave restricted files as world readable or writeable, a script changes the permissions to ensure that only members of the appropriate group are able to read or write to these files.
With the exception of users in ITAR or EAR groups, users are allowed to mount their ACCRE files to a local computer via SAMBA, which requires authentication. NFS mounting is not allowed since this method requires no user authentication.
THE BioVU SAMPLE REPOSITORY
BioVU, the Vanderbilt DNA Databank, is an enabling resource for exploration of the relationships among genetic variation, disease susceptibility, and variable drug responses, and represents a key first step in moving the emerging sciences of genomics and pharmacogenomics from research tools to clinical practice. A major goal of the resource is to generate datasets that incorporate de-identified information derived from medical records (See Synthetic Derivative (SD) described below) and genotype information to identify factors that affect disease susceptibility, disease progression, and/or drug response. The scale of the biobank is large (~259,000 samples as of spring 2021) Rapid accrual of samples is a result of Vanderbilt’s unique ability to de-identify the samples, which in part leads to a non-human subject designation with the Institutional Review Board, thus allowing the use of blood samples collected for clinical care and scheduled to be discarded. The program has received approval from the IRB and was reviewed in detail by the federal Office for Human Research Protections (OHRP), who agreed with the Non-Human Subjects regulatory designation for both the resource and subsequent research.
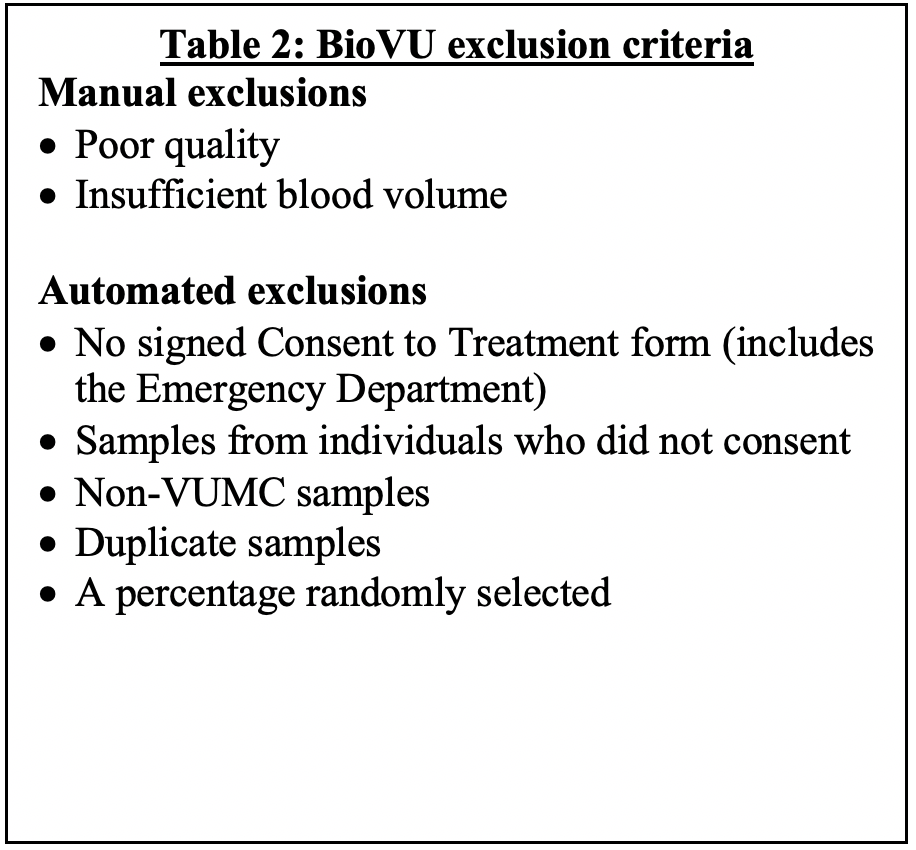
Program planning (e.g. Community and Ethics Committee involvement, sample handling routines) started in 2004, and sample accrual started at the end of February 2007. BioVU operates under a consent model for accumulation which is provided to patients in the clinic environments at Vanderbilt. The BioVU Consent form states policies on data sharing, privacy and should a signature be obtained, makes any leftover blood eligible for BioVU banking, subject to exclusions in Table 2.
Sample handling: BioVU has a comprehensive set of policies and procedures associated with the sample processing for acceptance (or rejection) into the repository. Sample processing includes prepping the blood specimens for scanning and the discard of some samples (based on visual inspection) due to low volume or quality issues; scanning samples through the custom-developed sample acceptance program (includes automated rejection of certain samples based on specific inclusion and exclusion criteria); creation of a record in the database for accepted samples using the hash algorithm described below; extraction of DNA from whole blood, discard of remaining blood products, quantitation of DNA; and long term storage of DNA samples in the Vanderbilt genomics core (VANTAGE) described below under appropriate storage conditions. The sample acceptance system consists of a series of software applications interacting with one another and with external software and hardware. Its role is to determine whether a given blood sample will be accepted into the biobank. The system has been tested and validated.
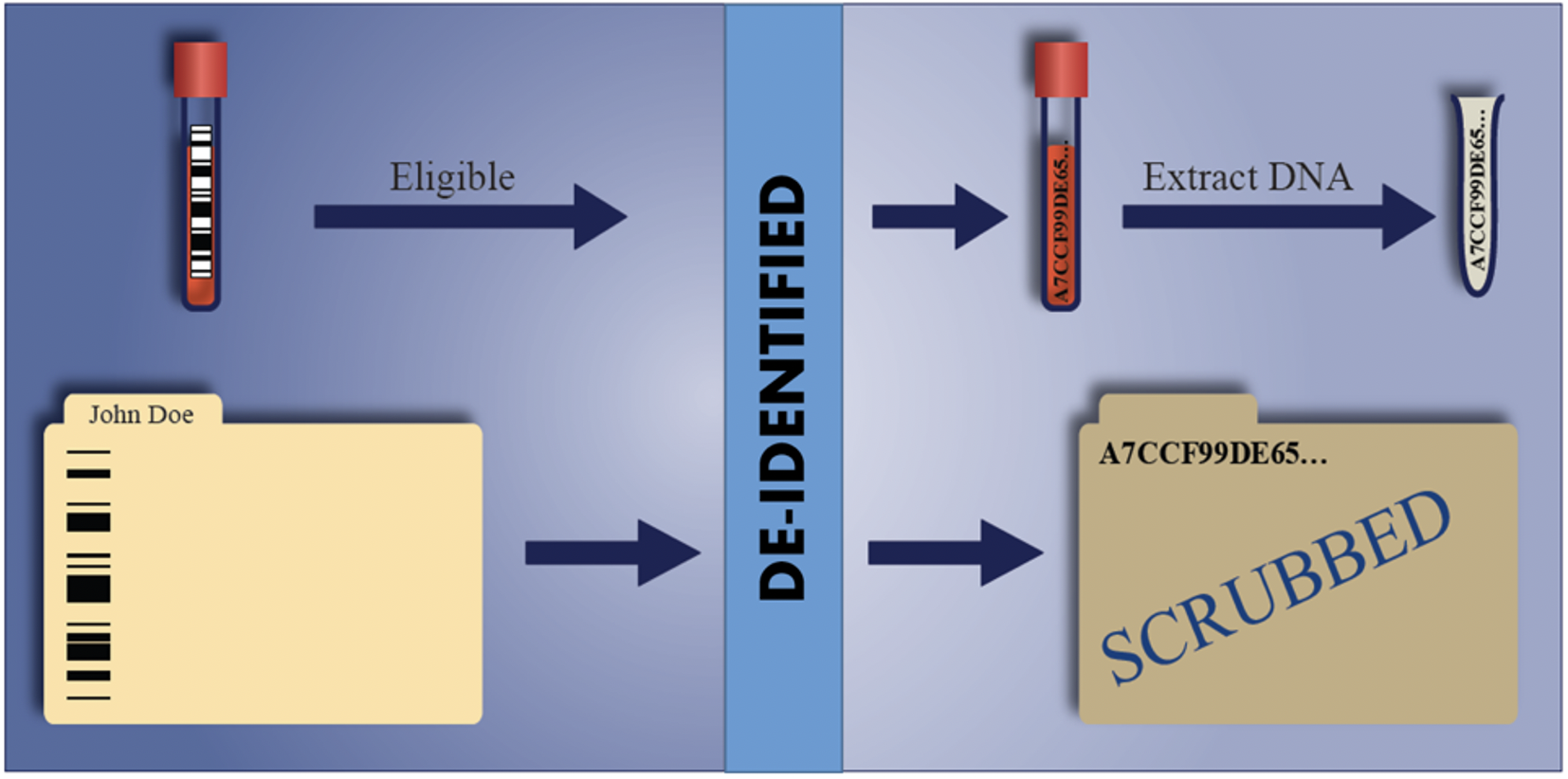
Once a sample passes all criteria, it is accepted by the program. Acceptance triggers the encryption program to assign a unique research ID number. The Secure Hash Algorithm (SHA) is a published and verified hash function employed for BioVU that has the property of generating a unique output for every unique input, and the property that the input cannot be inferred by cryptanalysis of the output. Using the Vanderbilt medical record number as an input, the system generates a unique, 512 bit (128 character) code that serves as a Research Unique Identifier (RUI) and is used to relate samples with de-identified computer data. We have validated that our version is performing to published encryption standards, i.e. it is not possible to infer or compute from the RUI the Medical Record Number that generated it (Figure 1).
DNA extraction and storage: DNA extraction relies on extraction protocols and systems currently utilized in the VANTAGE genomics resource. The Laboratory Information Management System (LIMS) tracks the samples throughout the extraction process. Data on the extraction process is stored in the LIMS-linked database and is available for review following extraction. The LIMS system records data on the extraction process (includes reagents used); initial blood volume (post extraction; the amount of sample actually stored); date, time, and type of extraction protocol; and run anomalies. Concentrations for each sample are quantitated fluorescently. Core personnel record the concentration information using the PTS TracerPlus program and upload these data to the LIMS database for each DNA sample. The sample acceptance system recognizes if there is <10 μg DNA left in storage and selectively accepts further blood samples from these individuals. If the banked sample contains >10 µg of DNA, further samples are not accepted for extraction. BioVU uses discarded blood samples collected during routine patient care, linked to de-identified data, extracted, and continuously updated from the electronic EMR.
 Sample characteristics and estimates: Samples accrued to date have a wide range of chronic and acute conditions, based on associated International Classification of Diseases ICD-9 or ICD-10 codes (which are extracted from the corresponding de-identified medical records). Of those currently collected samples, 43% are male and 57% female. The samples reflect the surrounding community, and are 77% from Caucasians and 9.76% African American. Asian and Hispanic populations comprise ~3.5%. Pediatric samples (individuals 0 to 18 years of age) comprise just over 10% of all BioVU samples.
Sample characteristics and estimates: Samples accrued to date have a wide range of chronic and acute conditions, based on associated International Classification of Diseases ICD-9 or ICD-10 codes (which are extracted from the corresponding de-identified medical records). Of those currently collected samples, 43% are male and 57% female. The samples reflect the surrounding community, and are 77% from Caucasians and 9.76% African American. Asian and Hispanic populations comprise ~3.5%. Pediatric samples (individuals 0 to 18 years of age) comprise just over 10% of all BioVU samples.
Existent genetic data: Genetic data derived from investigator-driven projects and institutional/program initiatives result in a nested in silico resource within the BioVU Databank. By signing the data use agreement, approved investigators are required to redeposit any genetic data derived on BioVU DNA samples back into the BioVU Databank. To date, more than 116,000 BioVU subjects have dense genetic data available for research (including ~108,000 subjects are at the Genome Wide Associated (GWA) level and >38,000 subjects have genotyping data associated from the ExomeChip) plus an additional large cohort with measured telomere length.
The “Synthetic Derivative” (SD). The Synthetic Derivative database is a research tool developed to enable studies with de-identified clinical data. The SD collection includes information extracted from the EMR systems described above, and indexed by the same one-way RUI used to track samples. Content is changed by deletion or permutation of all identifiers contained within each record, as described in the proposal.
The SD contains EMR data from >3.3 million total patients, with highly detailed longitudinal clinical data for approximately two million subjects, with an average clinical record size of 106,727 bytes (about 30 pages of text) and an average of 50 distinct diagnostic codes (ICD9 or ICD-10) per record. The database incorporates data from multiple sources and includes diagnostic and procedure codes (ICD-9 and Current Procedural Terminology [CPT]), basic demographics (age, gender, race), text from clinical care including discharge summaries, nursing notes, progress notes, history and physical, problem lists and multi-disciplinary assessments, laboratory values, ECG diagnoses, clinical text and electronically derived trace values, and inpatient medication orders. All clinical data are updated regularly to include patients new to VUMC, and therefore the SD, and to append new data to clinical records of existing patients as they continue to access care at VUMC. Thus, the resource is entirely suitable for mining information relative to disease progression over time.
The SD is stored on a secure server housed in the Vanderbilt Data Center and is fully compliant with the administrative, physical, and technical provisions of the HIPAA Security and Privacy Rules. The SD database is hosted on a Sun Solaris Server and running on Oracle 10g Software. The SD is mirrored by fully redundant server architecture in the event of a production server failure.
Queries. Simple queries to the Synthetic Derivative and BioVU resources (how many records meet certain criteria; how many have DNA samples) are available by a user interface hosted on the Vanderbilt University Medical Center intranet. Search criteria may include structured data fields such as ICD and CPT codes, demographics such as male or female, race and even specific age ranges. Search criteria includes the ability to search by specific medications and/or lab values, each of which can be used to include or exclude records returned. Unstructured text (i.e. free text) can be searched using specific keywords. With IRB approval, the user interface will also return data from individual subjects and provides text from the clinical record relative to specific keywords provided by the user. In this way, records can be included or excluded from further analysis. DNA samples corresponding to selected records can be requisitioned from the DNA Resources Core. Research using the records contained within the Synthetic Derivative can be initiated via a custom search interface after obtaining IRB approval.
CENTER FOR PRECISION MEDICINE PHEWAS CORE
About PheWAS. Methods to identify gene-disease associations primarily rely on clinical trials or observational cohorts and, more recently, Electronic Medical Record-linked DNA Biobanks. At Vanderbilt, we have used the EMR-linked DNA biobank BioVU to derive case and controls populations using data within the EMR to define clinical phenotypes. Genetic data for these EMR-linked association studies are redeposited into BioVU for future EMR-linked studies. This has opened the possibility of “reverse GWAS” or “Phenome-wide association studies” (PheWAS).
PheWAS using ICD9 codes. Our EMR-based PheWAS uses a custom-developed grouping of ICD-9 codes. These groupings loosely follow the 3-digit (category) and section groupings defined with the ICD-9 code system itself, but vary to include, for example, all hypertension codes (401-405) as one grouping. Each custom PheWAS code group also has an associated control group that excludes other related conditions (e.g., a patient with Graves disease cannot be a control for thyroiditis). Our original PheWAS in 2010 using ICD-9 codes replicated previously known gene-disease associations for 4/7 diseases (see publication) using records from BioVU, the Vanderbilt DNA biobank. Replicated associations included multiple sclerosis, rheumatoid arthritis, Crohn’s disease, and ischemic heart disease. The original PheWAS had 744 clinical case groups. A 2013 study using this revised model with 1645 hierarchically arranged phenotypes analyzed 3144 SNPs, replicated 210/751 associations (including 66% of those with adequate sample size), and noted 63 new, potentially pleiotropic associations. The PheWAS Core is updating this procedure to include ICD-10 codes. See http://phewascatalog.org, an online catalog of these results.
CENTER FOR PRECISION MEDICINE PRECISION PHENOTYPING CORE
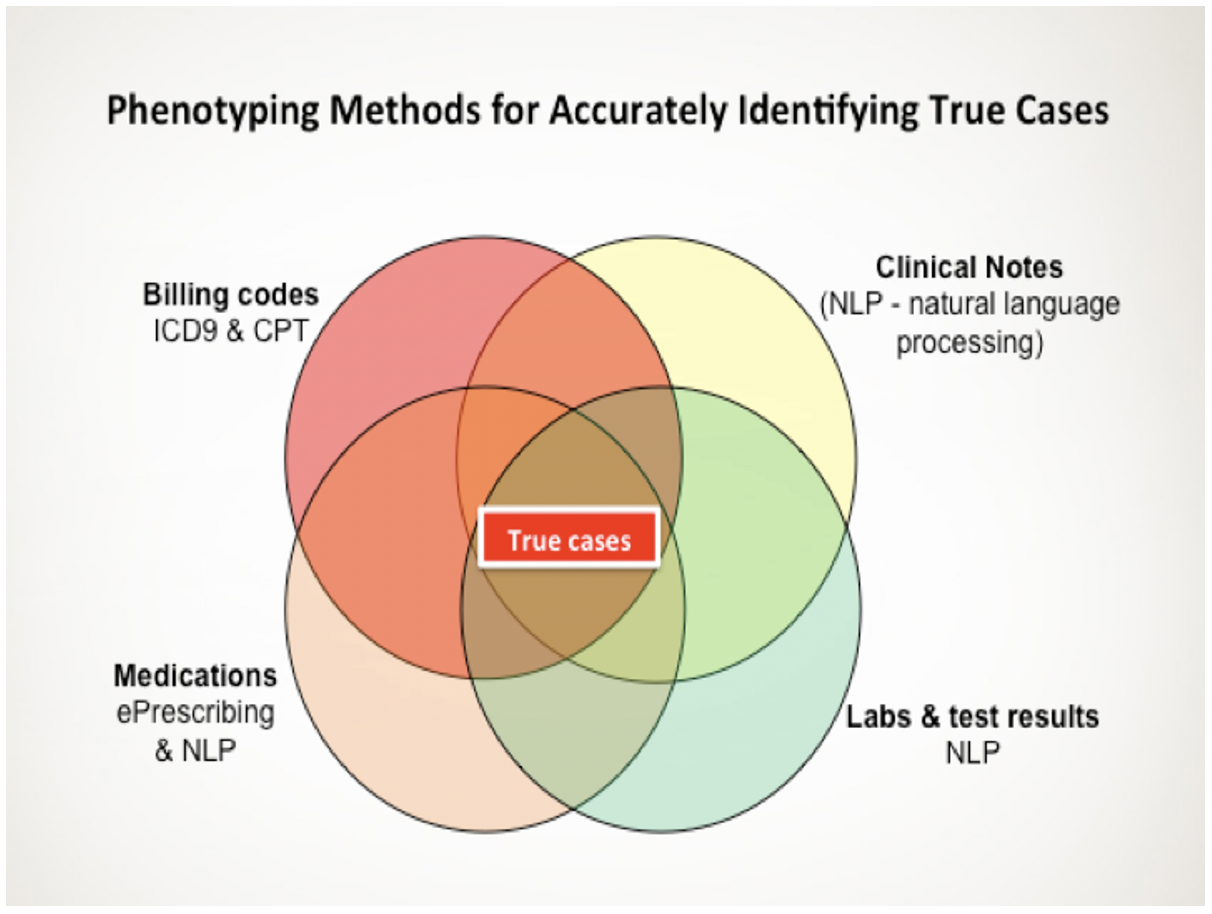 The mission of precision phenotyping core is to enable better research through better phenotyping. The core offers state-of-the-art informatics services to address the growing needs of the scientific community for Electronic Health Record (EHR) based phenotyping. They assist researchers who seek to advance the basic informatics methods, and to apply advanced methods to “understand” unstructured, and sometimes inaccurate, biomedical text and EHR data by providing services for clinical phenotype extraction, EHR-algorithm development, natural language processing methods, terminology/ontology reinforcement, machine learning, and integration of disparate data sources (such as coded and free text sources). For basic science applications, the core provides services for natural language processing methods, terminology development, and integration of disparate data sources (such as coded and free text sources).
The mission of precision phenotyping core is to enable better research through better phenotyping. The core offers state-of-the-art informatics services to address the growing needs of the scientific community for Electronic Health Record (EHR) based phenotyping. They assist researchers who seek to advance the basic informatics methods, and to apply advanced methods to “understand” unstructured, and sometimes inaccurate, biomedical text and EHR data by providing services for clinical phenotype extraction, EHR-algorithm development, natural language processing methods, terminology/ontology reinforcement, machine learning, and integration of disparate data sources (such as coded and free text sources). For basic science applications, the core provides services for natural language processing methods, terminology development, and integration of disparate data sources (such as coded and free text sources).
In conjunction with the PheWAS Core, the Precision Phenotyping Core provides two major domains of application of advanced informatics techniques: educational informatics and clinical phenotyping (deriving phenotypes from EMR records). The latter facilitates the discovery of clinical knowledge, gene-disease relationships (genomics, GWAS, and PheWAS), and gene-drug-outcome relationships (pharmacogenomics).
THE GENOME EDITING RESOURCE (Formerly: The Transgenic Mouse / ES Cell Shared Resource)
This Core currently or has previously (using the state-of-the-art technology available at the time) assisted investigators in generating, maintaining and storing germline-altered mice, through a wide array of services including CRISPR/Cas9 mutagenesis, gene targeting, pronuclear DNA microinjections, ES cell microinjection into blastocysts, assisted reproduction, BAC recombineering, and embryo cryopreservation and long-term storage. This Core played a central role in the generation of both the mice containing a germline G6pc2 deletion and mice containing a floxed G6pc2 allele that are described in the Research Strategy.
THE MOLECULAR CELL BIOLOGY RESOURCE
This Core facilitates bulk pricing for external synthesis of DNA oligonucleotides as well as the procurement of custom cell culture media, which will be used for the isolated islet and tissue culture studies, at a fraction of the cost of commercial sources. The supply arm of this resource stocks freshly prepared bacteria growth plates with or without antibiotics, transformation competent bacteria, and commercial kits and reagents for molecular biology research. It also houses instrumentation for imaging and quantitation of fluorescent, radioactive, luminescent, and chemiluminescent signals from gels, membranes, or multiple well plates.
THE CELL IMAGING SHARED RESOURCE
This Core provides access to a wide range of light, electron and digital imaging capabilities. The resource is staffed with individuals who provide instruction in the use of their many confocal microscopes, in how to perform immuno- and enzyme-cytochemistry, whole-mount microscopy, and image processing, pattern analysis, and 3-D reconstruction. Microscopes include a Zeiss LSM 710 inverted confocal microscope with climate-controlled environment for time-lapse experiments, an LSM 510 META inverted confocal microscope and a Zeiss Axiophot Microscope. In Aim 2 we propose using this core for the characterization of glucose-stimulated NAD(P)H generation in islets isolated from WT and G6pc2 KO mice.
VANTAGE (Vanderbilt Technologies for Advanced Genomics)
VANTAGE provides Vanderbilt researchers with guided access to a diverse and comprehensive set of genomics workflows, including next generation sequencing, gene expression, biobanking and genotyping. Occupying 12,500 square feet of renovated lab space, VANTAGE is supported by over $9M of federal and institutional funding. Dr. Simon Mallal serves as the Faculty Director of VANTAGE. With 25 years’ experience establishing and supervising high-throughput laboratories, Dr. Mallal has been instrumental in broadening VANTAGE’s NGS capacity. Dr. Mallal’s automation expertise combined with Associate Director Dr. Tom Stricker’s extensive experience in cancer genomics and NGS techniques and Deputy Director Dr. Suman Das’s microbial knowledge make VANTAGE a leader in cutting edge Genomic Research. Research, development, and technical aspects of VANTAGE are overseen by Angela Jones who has more than ten years of molecular biology experience with the last eight years focusing exclusively on high-throughput technologies and analyses. Project consultation, assay design, data management, and primary data analysis are supervised by Angela Jones. The core’s full-time operations manager, Karen Beeri, MS, manages the core’s day-to-day functions. Financial oversight is provided for VANTAGE and all VUMC research core resources by the Office of Research, under the direction of Jennifer Pietenpol PhD, Executive Vice President of Research of VUMC.
Sequencing, Expression, and Genotyping services. VANTAGE employs multiple robotic work stations in a well-equipped lab to conduct all phases of library preparation and quality control testing. Multiple Illumina NovaSeq 6000s and two MiSeqs provide high sequencing capacity and rapid turnaround time. VANTAGE users also have access to a full range of experimental approaches, including NanoString and 10X Genomics platforms, for gene expression studies down to the single-cell level. VANTAGE users also have access to a full range of experimental approaches for low- to high-throughput genotyping assays, including the Illumina array-based platform.
Extraction and Biobanking services. VANTAGE offers nucleic acid extraction, quantitative real-time PCR, and biobanking services by an experienced staff of technicians. The BioVU DNA Repository (Vanderbilt bio-bank, referenced above) is operated by and housed within VANTAGE. The Bio-bank workflow is automated, from whole blood extraction, to sample banking and subsequent pulls for robotic plating, with an integrated LIMS and sample database. The three-ton RTS SmaRTStore automated retrieval and storage system is supported by enhanced mechanical, electrical and plumbing infrastructures to ensure that pressure and humidity of compressed air, network connections, and correct electrical reach and delivery paths are provided. Security and accessibility to the robot room is limited to appropriate staff.
Advanced robotics from Tecan, Beckman Coulter, and Perkin Elmer support workflows for sample QC and labeling to hybridization, and data collection in our state-of-the-art lab. All VANTAGE service lines are supported by a modern LIMS for project submission, data delivery, and billing. Vanderbilt institutional assets for data management (Blue Arc) and data analysis (ACCRE) are backstopped by substantial VANTAGE computer resources.
VANGARD (Vanderbilt Technologies for Advanced Genomics Analysis and Research Design)
VANGARD is a core with administrative oversight from the Office of Research and scientific and technical direction provided by the Vanderbilt Center for Quantitative Sciences. The mission of VANGARD is to allow investigators to leverage the opportunities provided by next-generation sequencing and state-of-the-art genomics technologies. Offices, conference rooms, storage space, and computational space are located primarily on the 5th floor of the Preston Building within the Vanderbilt-Ingram Cancer Center. Dr. Yu Shyr serves as the director of VANGARD and maintains close communication with the leadership of VANTAGE to ensure seamless service delivery for genomic research. VANGARD operates in conjunction with VANTAGE, providing experimental design, quality assessment of data, quantitative genomic data analysis and results interpretation, and data storage. VANGARD also provides biostatistical and bioinformatic support for all genomic experiments that utilize BioVU specimens.
ZEBRAFISH AQUATIC FACILITY (Z-CORE)
The Zebrafish Aquatic Facility is composed of approximately 100 research tanks available for short and long-term studies, and a significant expansion project is underway. The facility enables investigator access to the unique advantages of the zebrafish model, which provides hundreds of mutant and transgenic zebrafish with unique properties suited for specific experiments. This core’s essential mission is to provide a reliable mechanism for incorporation and propagation of new and existing zebrafish lines into the core.
OTHER RESOURCES
Vanderbilt University Center for Teaching
The Vanderbilt University Center for Teaching promotes university teaching that leads to meaningful student learning. By helping members of the Vanderbilt community become more effective teachers, the Center for Teaching supports the educational mission of the university and enhances the learning experiences of its students.The Center offers programs and services in four overlapping areas: foundational teaching skills, new teaching practices, campus conversations on teaching and learning that are informed by national and international higher education developments, as well as local issues and priorities; and Identifying, sharing, and advocating for research-based practices in university teaching and the resources that support them.
The Center’s offerings are available to any members of the Vanderbilt community interested in developing their teaching practices. This benefits the HGEN program instructors as well as predoctoral students interested in developing their foundational teaching skills and exploring practices for a future in front of the classroom. The Center offers one-on-one consultation, seminars and facilitates retreats, curricula reviews, and other group activities.
Zerfoss Student Health Center
The Zerfoss Student Health Center serves the primary care needs of the Vanderbilt student community. There are no office co-pays for routine visits, and all registered students are eligible for care, regardless of insurance coverage. The Center’s mission enhances the academic experience of students by providing quality primary healthcare services in a nurturing and cost-effective manner. All of the physicians and nurse practitioners have chosen student health as their area of expertise and interest, and are dedicated to meeting the unique health care needs of the student population.
University Counseling Center (UCC)
The UCC supports the mental health needs of Vanderbilt students, encouraging their work toward their academic and personal goals. Their highly skilled and multidisciplinary staff develop evidence-based treatment plans tailored to each individual’s unique background and needs by working together with students, campus partners, and community providers. The UCC also emphasizes prevention and education through collaboration and consultation focused on the development of the skills and self-awareness needed to excel in a challenging educational environment. The Center’s philosophy is to offer culturally responsive services in a safe and affirming space with a commitment to ongoing dialogue. The UCC is committed to understanding the role of diverse experiences and backgrounds in order to best promote mental health and wellbeing in the lives of individuals and in our community.
The Annette and Irwin Eskind Biomedical Library and Informatics Center
Annette and Irwin Eskind Biomedical Library (EBL): this state-of-the-art library provides patrons access to information worldwide through the very latest in informatics retrieval and management technology, traditional library services, book stacks and comfortable reading areas are also provided along with technology training and assistance. The Research Informatics Consult Service (RICS), open to all members of the research community, provides proactive, targeted information services for Vanderbilt researchers delivered at the point of need. Services include training, grant assistance, electronic resources, database searching, bibliographic databases, full text resources, and molecular biology databases. Individual and group consultations are available with experienced information specialists. The library has a comprehensive, multidimensional Digital Library that offers fast, targeted access to online books, journals, databases and websites. EBL provides access to over 2,800 full-text electronic journal titles. EBL has developed proactive mechanisms to integrate evidence into clinical and research workflow through linkages of patient care guidelines within the electronic medical record. The library has a Patient Informatics Consult Service (PICS) program to provide patients/families with the latest health information: comprehensive information packets tailored to patient needs, access to the EBL’s consumer health materials collection and access to the EBL’s online Consumer Health Digital Library.
The VUMC/VUSM Office for Diversity Affairs, VUMC Office of Diversity and Inclusion, and the Vanderbilt University Vice Chancellor for Equity, Diversity and Inclusion
Both the University and Medical Center provide diversity, equity and inclusion resources and support to faculty, staff and students. The Vanderbilt University Medical Center/Vanderbilt University School of Medicine Office for Diversity Affairs is committed to the acceptance and graduation of a diverse group of trainees that is populated with members of all races, ethnicities, genders, gender identities, sexual orientation, disabilities, all places of geographic origin and the full spectrum of socio-economic status. The Office, led by VU Vice Chancellor for EDI Andre Churchwell, M.D., is also committed to and supportive of hiring faculty and staff who are equally diverse. The VUMC Office of Diversity and Inclusion is outward facing and provides faculty, staff and patients information about VUMC’s diversity and inclusion initiatives, as well as topical programs and resources. This office also supports the work of the Diversity Liaison Committee. The Vanderbilt University Vice Chancellor for Equity, Diversity and Inclusion (EDI) serves as an advisor in matters of diversity and inclusion for the Chancellor. The Vice Chancellor for EDI ensures that Vanderbilt’s seminal efforts are promulgated across the campus and the greater community. In addition, the Vice Chancellor for EDI’s focus will be on activities that advance diversity and inclusion tools and best practices with all Vice Chancellors and Vanderbilt University staff and in addition, serve when requested, as a supportive counsel to the Vice Provost of Strategic Initiatives/Office of Inclusive Excellence.
Health Information Privacy Laboratory (HIPLAB)
The Health Information Privacy Laboratory (HIPLAB) at Vanderbilt University, directed by Brad Malin, PhD, was founded to address the growing needs for privacy technology research and development for the emerging health information technologies sector. The goal of the HIPLAB is to improve the protection of patients’ privacy in health information systems. The HIPLAB is based in the Department of Biomedical Informatics, in the School of Medicine, and has relationships with various departments around the university and beyond. The HIPLAB performs basic, as well as, applied research in a number of health-related areas, including primary care and secondary sharing of patient-specific data for research purposes. Projects in the HIPLAB are multi-faceted and draw upon methodologies in computer science, the biomedical sciences, and public policy. The HIPLAB aims to improve the standard of healthcare and health information systems by developing technologies that enabling trust.