By Lorena Infante Lara
DNA replication is an intensely complicated process that relies on the actions and interactions of many, many proteins. At least 593, to be precise. Using iPOND (isolation of proteins on nascent DNA), a tool that was first developed in his lab, David Cortez (Biochemistry) has identified – and validated – over 590 proteins that are enriched in replication forks compared to the rest of the genome. His lab compiled this information into a resource that anyone can tap and published it in Cell Reports.
The research, spearheaded by postdoctoral fellow Sarah Wessel, answers a question that others have attempted in the past, albeit more fully than before.
“There is a significant lack of overlap between the previous resources and a lack of functional validation, making it difficult to assess how much of the proteomes is real and how much is noise,” said Wessel.
In the iPOND technique, the Cortez lab uses a thymidine analog, 5-ethynyl-2’-deoxyuridine (EdU), to label freshly made DNA. The label enables them to crosslink and isolate all proteins bound to the newly synthesized genetic material. The researchers can then identify which proteins are enriched or depleted at replication forks compared to the rest of the genome.
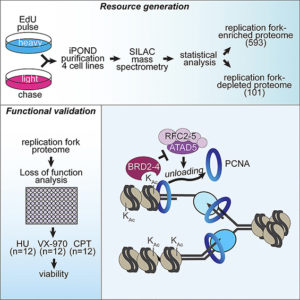
To construct the resource presented in this paper, Wessel and colleagues used iPOND to probe four different human cell lines. Eliminating “hits” that were not observed in at least two of the cell lines and adjusting for a 5% false discovery rate allowed them to whittle down an initial list of over 4000 different proteins to the 593 that they report were enriched (101 proteins were also found to be depleted at replication forks, meaning that their primary role is outside of DNA replication). Due to the high level of overlap between cell types (462 proteins were present in at least three cell lines), the authors concluded that nascent DNA-associated proteins are conserved across different human cell lineages.
A key aspect of this paper is that the authors validated their findings by generating 4 different siRNAs against each of the 593 enriched proteins and then using the siRNAs to analyze cells’ abilities to overcome different types of DNA replication stress in the absence of each protein. The results indicate which proteins are important for cell division and viability.
This screen, coupled with prior information from the literature, indicates that at least 504 of the identified proteins indeed function to promote faithful DNA replication.
“Over 110,000 data points were analyzed in these functional screens,” said Wessel. “The robustness of our approach provides a high degree of confidence in the resource and will allow other researchers in the field to confidently mine our data for their own investigations.”
Finally, to fully demonstrate the value of the resource in helping to identify how new or known proteins regulate different DNA replication processes, the Cortez lab selected three related proteins identified in this screen – BET proteins BRD2, BRD3, and BRD4 – and determined that they inhibit the unloading of PCNA from the chromatin through interactions with ATAD5. Although there were reports that BET proteins could interact with ATAD5 prior to this validation, the function of this interaction was not known.
Identifying new DNA replication-related functions for these three proteins is only the beginning. Wessel indicated that a large number of current cancer treatment target proteins were found in the screen, as well as other proteins associated with disease. This new compendium could help researchers further characterize these and other proteins of interest, and could help identify new potential targets for drug development.
This research was supported by the National Institutes of Health (grants GM116616, CA239161, CA009582, GM126646), the Breast Cancer Research Foundation, the Vanderbilt-Ingram Cancer Center, and by a Susan G. Komen fellowship.