by Cameron I. Cohen, graduate student in the Lauren Jackson lab
Traditional cryogenic electron microscopy relies on imaging proteins frozen in a monolayer of vitrified ice. Subsequent single particle analysis compiles images of the protein in different orientations and outputs a 3D reconstruction. Unfortunately, if proteins overlap or favor a specific orientation, a high-resolution structure cannot be determined. Cryogenic electron tomography addresses some of these issues by tilting the sample stage and acquiring images from multiple angles.
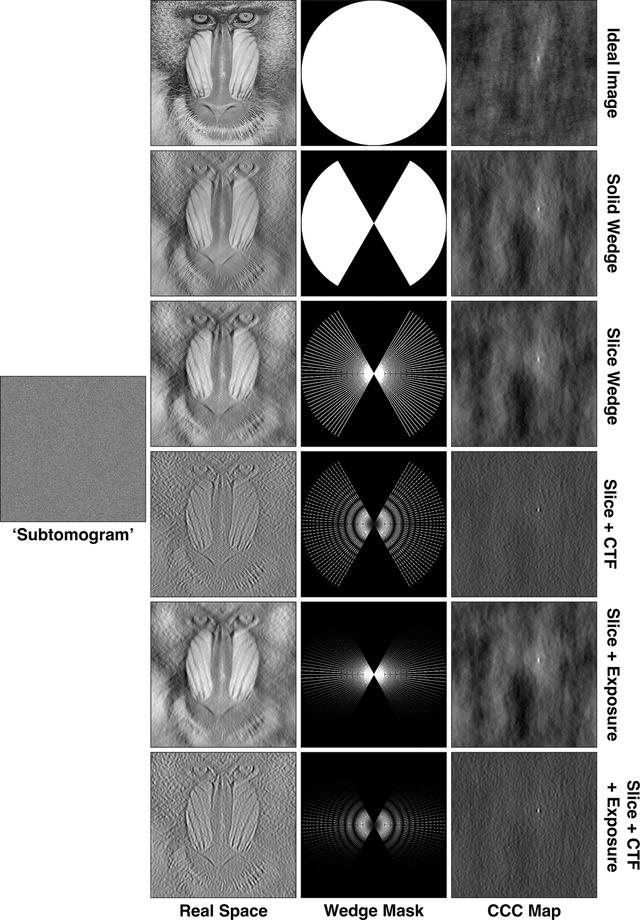
However, cryo-ET still suffers from the “missing wedge” problem. As samples become more tilted, their thickness at the extremes is increased and sample penetration decreases. Additionally, gaps between the angles imaged results in missing data. To address this problem, subtomogram averaging processes multiple tomograms—3D representations of the field of view—into a structure.

In this work, conducted by William Wan, assistant professor of biochemistry and affiliate of the Center for Structural Biology, and collaborators from the Max Planck Institute of Biochemistry, the authors describe the creation of a new STA package called STOPGAP.
STOPGAP is an open-source MATLAB package that uses a real-space correlation approach equipped with several new algorithms. Real-space correlation relies on the iterative construction of a reference image, calculation of correlation to the raw data, and refinement of the reference image. Distortions caused by the “missing wedge” can result in inaccurate processing, so STA employs a missing wedge filter to reference images. The missing wedge filter in STOPGAP is unique in that it reflects the sampling geometry and amplitude modulations present in tomographic data to ensure the reference image has the same aberrations as tomographic data. STOPGAP also employs a noise-correlation approach that subtracts a nonspecific correlation to increase the precision of subtomogram alignment. Finally, when images are sorted into different groups or “classes,” algorithms within the STOPGAP package encourage early switching to prevent subtomograms from becoming “trapped” in a class before distinct features emerge. This method will also produce a different result every time, allowing researchers to use consistency of classification as a marker for precision and accuracy.
This package not only improves several aspects of the current STA workflow, but it is also designed to fit within a user’s unique project needs. STOPGAP works with several existing cryo-ET packages, and the open-source nature of STOPGAP allows users fine-grained control over their data processing workflows.
To see if STOPGAP is right for your cryo-ET processing needs, read the full article in Acta Crystallographica Section D: Structural Biology!
This content was originally published in Weekly Coordinates, the newsletter of the Center for Structural Biology.