Small scale CRISPR screens in mice: iMAP, induced mosaic animals for perturbation
The Vanderbilt Genome Editing Resource develops emerging genome editing and modification technologies and offers access to Vanderbilt investigators through collaborative mechanisms. We are currently developing the production of arrays of sgRNAs that can be integrated into the mouse genome, then recombined by Cre-recombinase to produce cells of any tissue that contain single perturbations dependent upon the type of Cas9 expressed, as described in (Large-scale multiplexed mosaic CRISPR perturbation in the whole organism – PubMed (nih.gov)). We have recently produced our first mouse containing an array of six guides and experimental validation will soon be underway. We are looking for Vanderbilt investigators who may be interested in performing small scale screening in a living mouse to help us develop this emerging technology. Specific advantages:
- Targeted Perturbation: Unlike traditional CRISPR screening in cell lines, iMAP allows us to identify genes involved in critical biological processes within a living mouse.
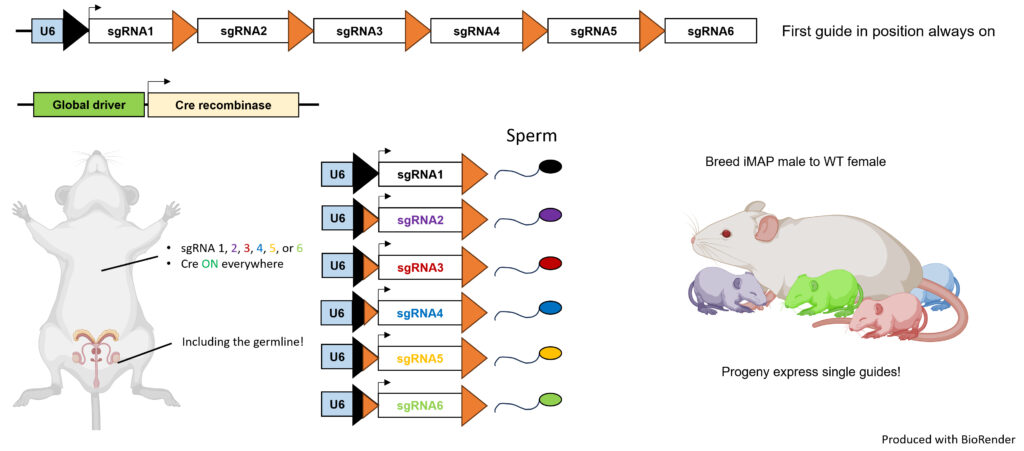
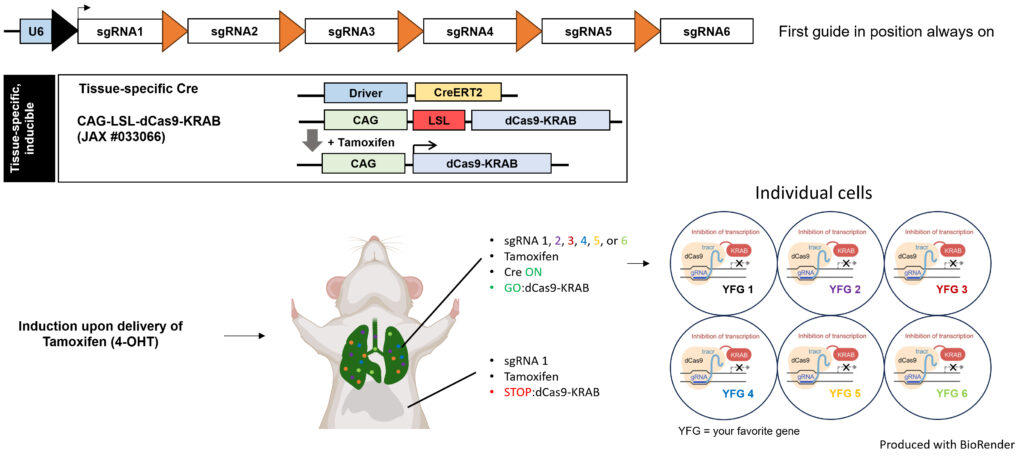
- Guide RNA Arrays: The iMAP system features guide RNA (sgRNA) expression arrays. These arrays utilize a single U6 promoter, with individual guide RNAs separated by mutant loxP sequences. When Cre is expressed, these sequences randomly recombine, ensuring that only one guide RNA is active per cell.
- Diverse Genetic Perturbations: Each cell will have a distinct genetic manipulation. Depending on the type of Cas9 expressed, we can achieve gene deletions, repression, or activation.
- Tissue Agnostic: With all elements genetically expressed, iMAP works in any tissue or cell type.
- Breed Mutations of Interest: iMAP arrays can be recombined in the germline with an appropriate Cre driver. Mice expressing individual sgRNAs may be bred to produce single perturbation progeny.
In vivo screening
Produce mice with single perturbations by breeding