Brandt Eichman and Walter Chazin, professors of biochemistry, have worked together to provide a better understanding of how exactly DNA replication is initiated in eukaryotes. Using Vanderbilt’s state-of-the-art instrumentation in the Center for Structural Biology’s Cryo-Electron Microscopy Facility, Eichman, Chazin, and their colleagues provided detailed visualizations of a multi-functional protein in action, which sheds light on how DNA replication is initiated in humans.
Eichman and Chazin shared reflections on this research, newly published in Nature Structural & Molecular Biology:
What issue does your research address?
We are interested in the molecular details of human DNA replication, one of the most fundamental processes of life; it is repeated millions of times each day as we make new cells. The new copies of DNA are synthesized by polymerases, which read the sequence of an existing DNA strand one nucleotide at a time and add the complementary nucleotide to the nascent DNA strand. Specific polymerases perform the bulk of DNA synthesis, but they are unable to function without first having a short “primer” segment of the new strand.
This work addresses the molecular mechanisms of DNA polymerase α–primase (polα–primase), the enzyme responsible for synthesizing the primers. Polα–primase is an essential enzyme as it is the only polymerase that can initiate DNA synthesis by generating the primers that the other polymerases need for duplication of the genome.
Despite polα–primase being the first human polymerase discovered, the way it synthesizes very specific lengths of RNA and DNA in a single strand remained unclear for more than fifty years. How does it know that it has synthesized a specific number ofnucleotides of RNA before transitioning to DNA synthesis? How does it transition between the two modes? How does it know that it has synthesized a certain number of nucleotides of DNA before stopping?
Understanding the mechanisms behind polα–primase’s ability to “count” the length of the RNA and DNA segments of the primer is important because primers must be kept to a very short length, as they contain RNA in the new DNA strand and the DNA synthesized by polα is littered with mutations. Thus, the primers would be highly detrimental to the cell if they became a substantial part of the new DNA strand that persisted in the genome after replication.
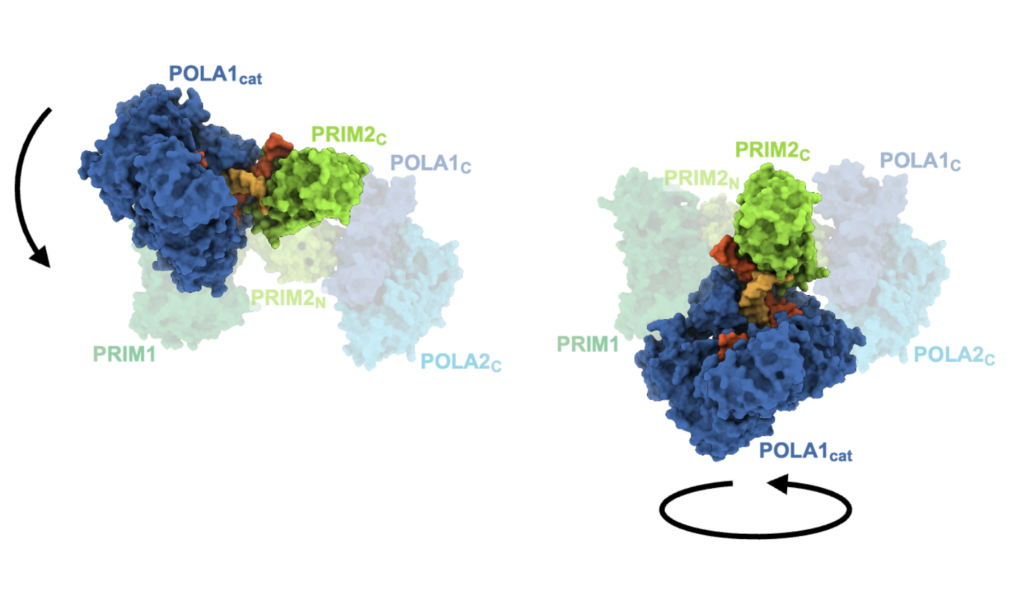
To answer these outstanding questions, we used cryo-electron microscopy to capture snapshots of this multi-functional protein at various stages as it generates a primer. The high-resolution structures we determined illuminated the mechanisms of RNA and DNA counting by polα–primase. They also provide a starting point for design of novel small molecule modulators of polα–primase function that would provide new ways to investigate DNA replication in cells.
What was unique about your approach to the research?
The Eichman and Chazin labs have collaborated for many years to understand how polα–primase works. We visualized some of the first structures of polα–primase bound to nucleic acid substrates. It was the highly strategic design of primer/template substrates that allowed our team to “trap” the enzyme at several specific points along the pathway to synthesizing the primer. Importantly, this research was made possible by access to the state-of-the-art instrumentation in the CSB Cryo-Electron Microscopy Facility.
What were your findings?
Our data directly show that polα–primase holds on to one end of the primer throughout all stages of synthesis. This observation is critical to understanding how the initial RNA-primed template is handed off from the primase active site in one subunit (where RNA synthesis occurs) to the DNA polymerase active site in another subunit (where DNA synthesis occurs). The sustained attachment also serves to increase polα–primase’s ability to remain bound to the template and to regulate both RNA and DNA composition. Importantly, the detailed analysis of the structures revealed how flexibility within this four-subunit complex is critical to being able to synthesize the primer strand across two active sites.
In addition, our research suggests that termination of DNA synthesis is facilitated by reduction polα and primase affinities for the template as more DNA is synthesized.
What do you hope will be achieved with the research results?
We hope our research findings will illuminate to the field a more complete understanding of replication initiation and contribute to the growing understanding that complex molecular machinery requires flexibility and dynamics to function. The inherent flexibility within this complex, multi-subunit polymerase is essential to primer synthesis and to its ability to dynamically interact with multiple other enzymes present in the replisome (for the handoff of the primer to the replicative polymerase for bulk DNA synthesis, for example).
We also hope that this work will lead to a better understanding of how current polα–primase inhibitors work and more broadly pave the way for future designs of small molecule modulators to serve as tools for studying DNA replication in cells. Tool compounds of this type can also be used to evaluate the therapeutic potential of targeting specific replication proteins with roles in diseases of genome instability.
GO DEEPER
The paper “A mechanistic model of primer synthesis from catalytic structures of DNA polymerase α–primase” was published in Nature Structural & Molecular Biology on March 15, 2024.
FUNDING
This research was funded by the National Institutes of Health.