MSTPublications: April 2019
 Respiratory Heterogeneity Shapes Biofilm Formation and Host Colonization in Uropathogenic Escherichia coli.
Respiratory Heterogeneity Shapes Biofilm Formation and Host Colonization in Uropathogenic Escherichia coli.
Beebout CJ, Eberly AR, Werby SH, Reasoner SA, Brannon JR, De S, Fitzgerald MJ, Huggins MM, Clayton DB, Cegelski L, Hadjifrangiskou M.
MBio. 2019 Apr 2;10(2). pii: e02400-18. doi: 10.1128/mBio.02400-18.
Biofilms are multicellular bacterial communities encased in a self-secreted extracellular matrix comprised of polysaccharides, proteinaceous fibers, and DNA. Organization of these components lends spatial organization to the biofilm community such that biofilm residents can benefit from the production of common goods while being protected from exogenous insults. Spatial organization is driven by the presence of chemical gradients, such as oxygen. Here we show that two quinol oxidases found in Escherichia coli and other bacteria organize along the biofilm oxygen gradient and that this spatially coordinated expression controls architectural integrity. Cytochrome bd, a high-affinity quinol oxidase required for aerobic respiration under hypoxic conditions, is the most abundantly expressed respiratory complex in the biofilm community. Depletion of the cytochrome bd-expressing subpopulation compromises biofilm complexity by reducing the abundance of secreted extracellular matrix as well as increasing cellular sensitivity to exogenous stresses. Interrogation of the distribution of quinol oxidases in the planktonic state revealed that ∼15% of the population expresses cytochrome bd at atmospheric oxygen concentration, and this population dominates during acute urinary tract infection. These data point toward a bet-hedging mechanism in which heterogeneous expression of respiratory complexes ensures respiratory plasticity of E. coli across diverse host niches. IMPORTANCE Biofilms are multicellular bacterial communities encased in a self-secreted extracellular matrix comprised of polysaccharides, proteinaceous fibers, and DNA. Organization of these components lends spatial organization in the biofilm community. Here we demonstrate that oxygen gradients in uropathogenic Escherichia coli (UPEC) biofilms lead to spatially distinct expression programs for quinol oxidases-components of the terminal electron transport chain. Our studies reveal that the cytochrome bd-expressing subpopulation is critical for biofilm development and matrix production. In addition, we show that quinol oxidases are heterogeneously expressed in planktonic populations and that this respiratory heterogeneity provides a fitness advantage during infection. These studies define the contributions of quinol oxidases to biofilm physiology and suggest the presence of respiratory bet-hedging behavior in UPEC.
 Broad domains of histone 3 lysine 4 trimethylation are associated with transcriptional activation in CA1 neurons of the hippocampus during memory formation.
Broad domains of histone 3 lysine 4 trimethylation are associated with transcriptional activation in CA1 neurons of the hippocampus during memory formation.
Collins BE, David Sweatt J, Greer CB.
Neurobiol Learn Mem. 2019 Apr 16. pii: S1074-7427(19)30075-9. doi: 10.1016/j.nlm.2019.04.009. [Epub ahead of print]
Transcriptional changes in the hippocampus are required for memory formation, and these changes are regulated by numerous post-translational modifications of chromatin-associated proteins. One of the epigenetic marks that has been implicated in memory formation is histone 3 lysine 4 trimethylation (H3K4me3), and this modification is found at the promoters of actively transcribed genes. The total levels of H3K4me3 are increased in the CA1 region of the hippocampus during memory formation, and genetic perturbation of the K4 methyltransferases and demethylases interferes with forming memories. Previous chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) analyses failed to detect changes in H3K4me3 levels at the promoters of memory-linked genes. Since the breadth of H3K4me3 marks was recently reported to be associated with the transcriptional outcome of a gene, we re-analyzed H3K4me3 ChIP-seq data sets to identify the role of H3K4me3 broad domains in CA1 neurons, as well as identify differences in breadth that occur during contextual fear conditioning. We found that, under baseline conditions, broad H3K4me3 peaks mark important learning and memory genes and are often regulated by super-enhancers. The peaks at many learning-associated genes become broader during novel environment exposure and memory formation. Furthermore, the important learning- and memory-associated lysine methyltransferases, Kmt2a and Kmt2b, are involved in maintaining H3K4me3 peak width. Our findings highlight the importance of analyzing H3K4me3 peak shape, and demonstrate that breadth of H3K4me3 marks in neurons of the hippocampus is regulated during memory formation.
Ben Brown’s recent first-author paper was featured here in the VUMC Reporter!
 Merla Hubler’s first-author paper was honored as the 2018 American Journal of Physiology paper of the year!
Merla Hubler’s first-author paper was honored as the 2018 American Journal of Physiology paper of the year!
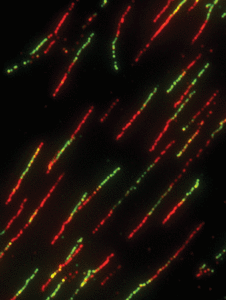
 Matthew Puccetti’s recent first-author paper was featured on the cover of Cancer Research! The image is a photograph of strands of DNA from primary mouse B cells isolated from Zranb3– or Smarcal1-deficient Eμ-myc transgenic mice pulse-labeled with IdU (red) and CldU (green) detected by immunofluorescence and microscopy. The technique is called DNA fiber labeling.
Matthew Puccetti’s recent first-author paper was featured on the cover of Cancer Research! The image is a photograph of strands of DNA from primary mouse B cells isolated from Zranb3– or Smarcal1-deficient Eμ-myc transgenic mice pulse-labeled with IdU (red) and CldU (green) detected by immunofluorescence and microscopy. The technique is called DNA fiber labeling.
MyD88 and IL-1R signaling drive antibacterial immunity and osteoclast-driven bone loss during Staphylococcus aureus osteomyelitis.
Putnam NE, Fulbright LE, Curry JM, Ford CA, Petronglo JR, Hendrix AS, Cassat JE.
PLoS Pathog. 2019 Apr 12;15(4):e1007744. doi: 10.1371/journal.ppat.1007744. [Epub ahead of print]
Predictors of endoscopic third ventriculostomy ostomy status in patients who experience failure of endoscopic third ventriculostomy with choroid plexus cauterization.
Hale AT, Stanton AN, Zhao S, Haji F, Gannon SR, Arynchyna A, Wellons JC, Rocque BG, Naftel RP.
J Neurosurg Pediatr. 2019 Apr 19:1-6. doi: 10.3171/2019.2.PEDS18743. [Epub ahead of print]
Imaging mass spectrometry enables molecular profiling of mouse and human pancreatic tissue.
Prentice BM, Hart NJ, Phillips N, Haliyur R, Judd A, Armandala R, Spraggins JM, Lowe CL, Boyd KL, Stein RW, Wright CV, Norris JL, Powers AC, Brissova M, Caprioli RM.
Diabetologia. 2019 Apr 6. doi: 10.1007/s00125-019-4855-8. [Epub ahead of print]
Sex modifies placental gene expression in response to metabolic and inflammatory stress.
Barke TL, Money KM, Du L, Serezani A, Gannon M, Mirnics K, Aronoff DM.
Placenta. 2019 Mar;78:1-9. doi: 10.1016/j.placenta.2019.02.008. Epub 2019 Feb 22.
The patient-independent human iPSC model – a new tool for rapid determination of genetic variant pathogenicity in long QT syndrome.
Chavali NV, Kryshtal DO, Parikh SS, Wang L, Glazer AM, Blackwell DJ, Kroncke BM, Shoemaker MB, Knollmann BC.
Heart Rhythm. 2019 Apr 17. pii: S1547-5271(19)30360-1. doi: 10.1016/j.hrthm.2019.04.031. [Epub ahead of print]